2N24
 
 | |
2N2G
 
 | |
2IUE
 
 | |
2MS1
 
 | |
2MQV
 
 | |
2MQT
 
 | |
2N19
 
 | | STIL binding to the Polo-box domain 3 of PLK4 regulates centriole duplication | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Boehm, R, Arquint, C, Gabryjonczyk, A, Imseng, S, Sauer, E, Hiller, S, Nigg, E, Maier, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | STIL binding to Polo-box 3 of PLK4 regulates centriole duplication.
Elife, 4, 2015
|
|
2NBA
 
 | |
2ILX
 
 | |
2N90
 
 | |
2NB4
 
 | | Solution structure of Q388A3 PDZ domain | | Descriptor: | Putative uncharacterized protein | | Authors: | Mei, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
2NPR
 
 | | Structural Studies on Plasmodium vivax Merozoite Surface Protein-1 | | Descriptor: | Merozoite surface protein 1 | | Authors: | Babon, J.J, Morgan, W.D, Kelly, G, Eccleston, J.F, Feeney, J, Holder, A.A. | | Deposit date: | 2006-10-29 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural studies on Plasmodium vivax merozoite surface protein-1
Mol.Biochem.Parasitol., 153, 2007
|
|
1WA7
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE IN COMPLEX WITH A HERPESVIRAL LIGAND | | Descriptor: | HYPOTHETICAL 28.7 KDA PROTEIN IN DHFR 3'REGION (ORF1), TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-10-25 | | Release date: | 2005-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck.
Biochemistry, 41, 2002
|
|
2K3N
 
 | |
2BW2
 
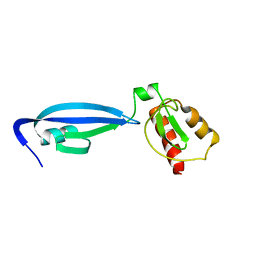 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
2K3O
 
 | |
2BTG
 
 | |
2MSA
 
 | |
2MQU
 
 | | Spatial structure of Hm-3, a membrane-active spider toxin affecting sodium channels | | Descriptor: | Neurotoxin Hm-3 | | Authors: | Myshkin, M.Y, Shenkarev, Z.O, Paramonov, A.S, Berkut, A.A, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2014-06-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of membrane-active toxin from crab spider Heriaeus melloteei suggests parallel evolution of sodium channel gating modifiers in Araneomorphae and Mygalomorphae.
J. Biol. Chem., 290, 2015
|
|
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2NUZ
 
 | | crystal structure of alpha spectrin SH3 domain measured at room temperature | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Agarwal, V, Faelber, K, Hologne, M, Chevelkov, V, Oschkinat, H, Diehl, A, Reif, B. | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of MAS solid-state NMR and X-ray crystallographic data in the analysis of protein dynamics in the solid state
To be Published
|
|
2EJY
 
 | |
2KM2
 
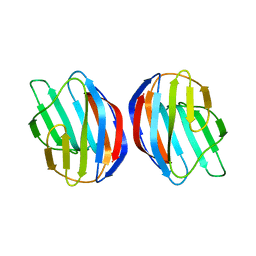 | | Galectin-1 dimer | | Descriptor: | Galectin-1 | | Authors: | Nesmelova, I.V, Ermakova, E, Daragan, V.A, Pang, M, Baum, L.G, Mayo, K.H. | | Deposit date: | 2009-07-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity.
J.Mol.Biol., 397, 2010
|
|
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | Descriptor: | PROTEIN L | | Authors: | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | Deposit date: | 1994-08-12 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
2I3E
 
 | |
