7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7OI0
 
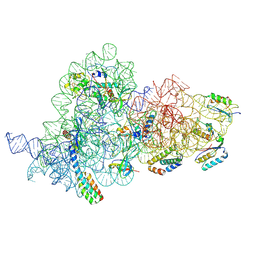 | | E.coli delta rbfA pre-30S ribosomal subunit class D | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Maksimova, E, Korepanov, A, Baymukhametov, T, Kravchenko, O, Stolboushkina, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center.
Int J Mol Sci, 22, 2021
|
|
6CVM
 
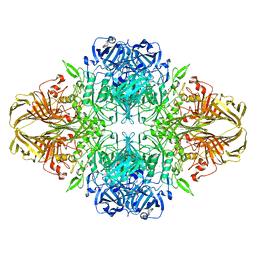 | | Atomic resolution cryo-EM structure of beta-galactosidase | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
5NWY
 
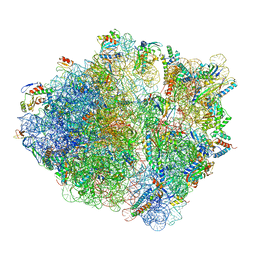 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
5NWS
 
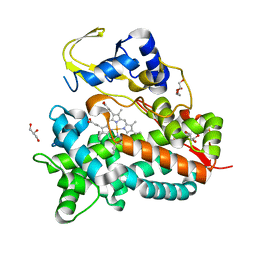 | | Crystal structure of saAcmM involved in actinomycin biosynthesis | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Driller, R, Semsary, S, Crnovicic, I, Vater, J, Keller, U, Loll, B. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Ketonization of Proline Residues in the Peptide Chains of Actinomycins by a 4-Oxoproline Synthase.
Chembiochem, 19, 2018
|
|
1UIT
 
 | | Solution structure of RSGI RUH-006, The third PDZ domain OF hDlg5 (KIAA0583) protein [Homo sapiens] | | Descriptor: | HUMAN DISCS LARGE 5 PROTEIN | | Authors: | Abe, T, Hirota, H, Kobayashi, N, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-22 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-006, The third PDZ domain OF hDlg5 (KIAA0583) protein [Homo sapiens]
To be Published
|
|
1UIP
 
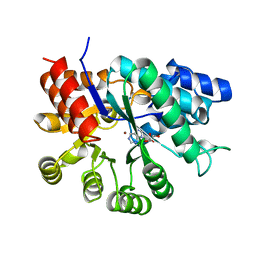 | | ADENOSINE DEAMINASE (HIS 238 GLU MUTANT) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
7TXQ
 
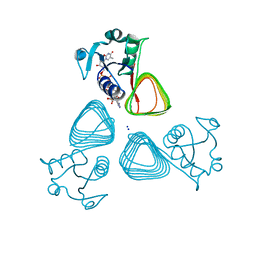 | | x-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in the present of TDP and Acetyl-CoenzymeA | | Descriptor: | ACETYL COENZYME *A, SODIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7TXS
 
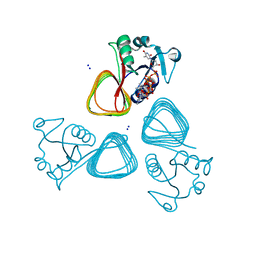 | | X-ray structure of the VioB N-aetyltransferase from Acinetobacter baumannii in the presence of a reaction intermediate | | Descriptor: | SODIUM ION, VioB, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (3R)-4-({3-[(2-{[(1S)-1-{[(2R,3S,4S,5R,6R)-4,5-dihydroxy-6-{[(R)-hydroxy{[(R)-hydroxy{[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)oxolan-2-yl]methoxy}phosphoryl]oxy}phosphoryl]oxy}-2-methyloxan-3-yl]amino}ethyl]sulfanyl}ethyl)amino]-3-oxopropyl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate (non-preferred name) | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
1V6X
 
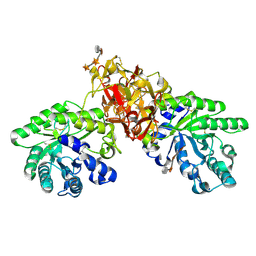 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1UJD
 
 | | Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA | | Descriptor: | KIAA0559 protein | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA
To be published
|
|
5FFO
 
 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
2CHD
 
 | | Crystal structure of the C2A domain of Rabphilin-3A | | Descriptor: | GLYCEROL, RABPHILIN-3A | | Authors: | Biadene, M, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the C2A Domain of Rabphilin-3A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7TXP
 
 | | X-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in complex with TDP-4-amino-4,6-dideoxy-D-glucose | | Descriptor: | SODIUM ION, VioB, dTDP-4-amino-4,6-dideoxyglucose | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
5OJ3
 
 | | YCF48 from Cyanidioschyzon merolae | | Descriptor: | Photosystem II stability/assembly factor HCF136 | | Authors: | Michoux, F, Murray, J.W, Nixon, P.J. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D2T
 
 | | HLA-B*57:01 presenting LALLTGVRW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
5DQT
 
 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5IKV
 
 | | The Structure of Flufenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
2CFS
 
 | |
5DYS
 
 | | Crystal Structure of T94I rhodopsin mutant | | Descriptor: | ACETATE ION, PALMITIC ACID, RETINAL, ... | | Authors: | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | Deposit date: | 2015-09-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
1XJL
 
 | |
2CFT
 
 | |
1R4E
 
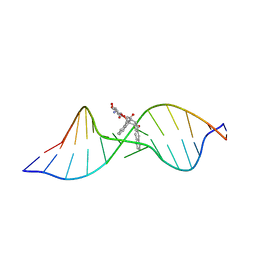 | | Solution structure of the Complex Formed between a Left-Handed Wedge-Shaped Spirocyclic Molecule and Bulged DNA | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*AP*GP*TP*TP*CP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*GP*AP*TP*GP*CP*GP*TP*G)-3', SPIRO[NAPHTHALENE-2(3H),3'(10'H)-PENTALENO[1,2-B]NAPHTHALENE]-3,10'-DIONE, ... | | Authors: | Hwang, G.S, Jones, G.B, Goldberg, I.H. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Stereochemical control of small molecule binding to bulged DNA: comparison of structures of spirocyclic enantiomer-bulged DNA complexes.
Biochemistry, 43, 2004
|
|
7OI9
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 3B | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
