7Z0Z
 
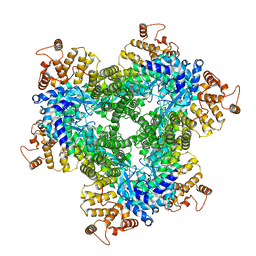 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis, Y44F variant | | Descriptor: | AbiK | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
4CMQ
 
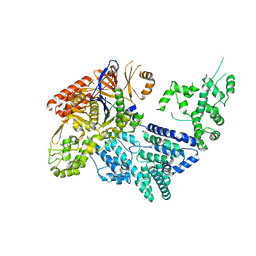 | | Crystal structure of Mn-bound S.pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MANGANESE (II) ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA- Mediated Conformational Activation
Science, 343, 2014
|
|
3LZ8
 
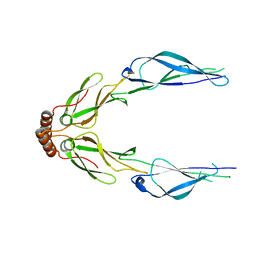 | | Structure of a putative chaperone dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.9 a resolution. | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Putative Chaperone Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 at 2.9 A Resolution.
To be Published
|
|
5CKI
 
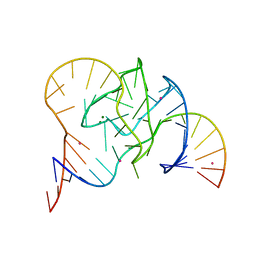 | | Crystal structure of 9DB1* deoxyribozyme (Cobalt hexammine soaked crystals) | | Descriptor: | COBALT (II) ION, DNA (44-MER), MAGNESIUM ION, ... | | Authors: | Ponce-Salvatierra, A, Hoebartner, C, Pena, V. | | Deposit date: | 2015-07-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of a DNA catalyst.
Nature, 529, 2016
|
|
2IQC
 
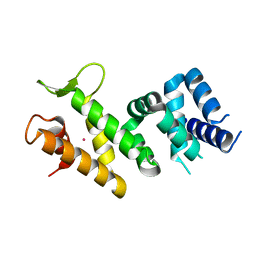 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
5JA4
 
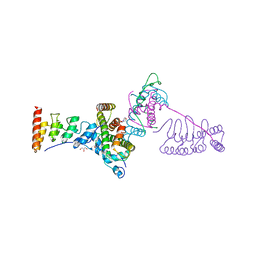 | |
7QWV
 
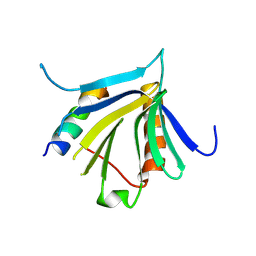 | | Crystal structure of the REC114-TOPOVIBL complex. | | Descriptor: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | Authors: | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
1PYJ
 
 | | Solution Structure of an O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct in an 11mer DNA duplex | | Descriptor: | 5'-D(*CP*CP*AP*TP*AP*TP*GP*GP*CP*CP*C)-3', 5'-D*GP*GP*GP*CP*CP*AP*TP*AP*TP*GP*G)-3' | | Authors: | Peterson, L.A, Vu, C, Hingerty, B.E, Broyde, S, Cosman, M. | | Deposit date: | 2003-07-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct in an 11 mer DNA duplex: evidence for formation of a base triplex.
Biochemistry, 42, 2003
|
|
1JO2
 
 | |
1XSL
 
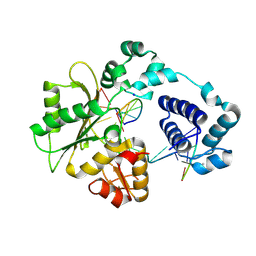 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap | | Descriptor: | 5'-D(*CP*GP*GP*CP*AP*GP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
6Q6R
 
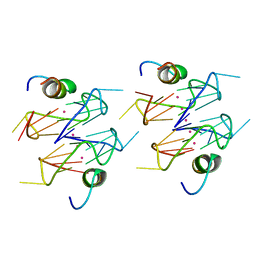 | | Recognition of different base tetrads by RHAU: X-ray crystal structure of G4 recognition motif bound to the 3-end tetrad of a DNA G-quadruplex | | Descriptor: | ATP-dependent DNA/RNA helicase DHX36, POTASSIUM ION, Parallel stranded DNA G-quadruplex | | Authors: | Heddi, B, Cheong, V.V, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3'-end tetrad of a DNA G-quadruplex.
J.Struct.Biol., 209, 2020
|
|
2VS8
 
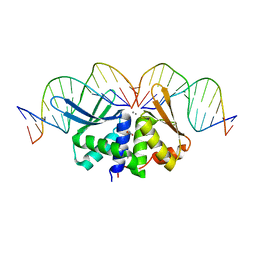 | | The crystal structure of I-DmoI in complex with DNA and Mn | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP* GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*A)-3', ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1N5C
 
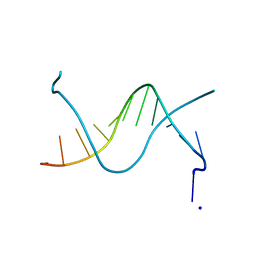 | | Crystal Structure Analysis of the B-DNA Dodecamer CGCGAATT(ethenoC)GCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDC)P*GP*CP*G)-3', SODIUM ION | | Authors: | Freisinger, E, Fernandes, A, Grollman, A.P, Kisker, C.F. | | Deposit date: | 2002-11-05 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic Characterization of an Exocyclic DNA Adduct: 3,N4-etheno-2'-deoxycytidine in the Dodecamer 5'-CGCGAATT(ethenoC)GCG-3'
J.Mol.Biol., 329, 2003
|
|
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
7ARQ
 
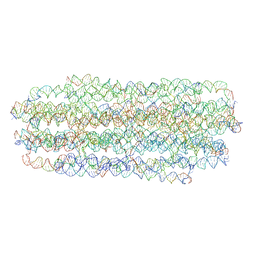 | | Cryo EM of 3D DNA origami 16 helix bundle | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
1XSN
 
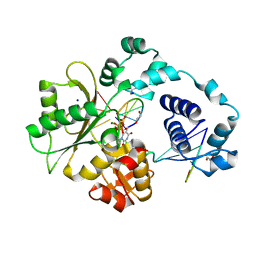 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap and ddTTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4CMP
 
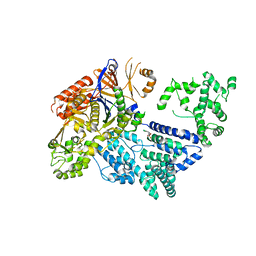 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
1P4Z
 
 | |
1XSP
 
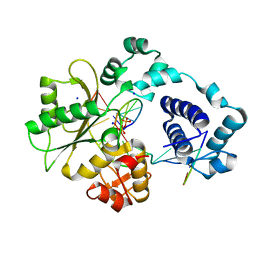 | | Crystal Structure of human DNA polymerase lambda in complex with nicked DNA and pyrophosphate | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1QDA
 
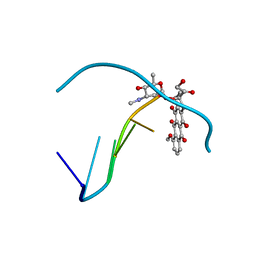 | | Crystal structure of epidoxorubicin-formaldehyde virtual crosslink of DNA | | Descriptor: | 4'-EPIDOXORUBICIN, 5' -D(CP*GP*CP*(G49)P*CP*GP)-3' | | Authors: | Podell, E.R, Harrington, D.J, Taatjes, D.J, Koch, T.H. | | Deposit date: | 1999-05-17 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of epidoxorubicin-formaldehyde virtual crosslink of DNA and evidence for its formation in human breast-cancer cells.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4EUG
 
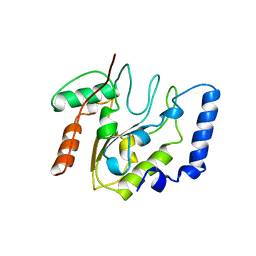 | | Crystallographic and Enzymatic Studies of an Active Site Variant H187Q of Escherichia Coli Uracil DNA Glycosylase: Crystal Structures of Mutant H187Q and its Uracil Complex | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187.
Biochemistry, 38, 1999
|
|
1TF3
 
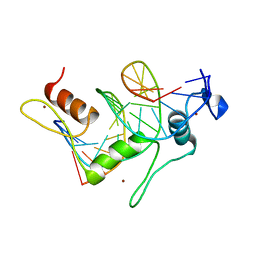 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
4EFJ
 
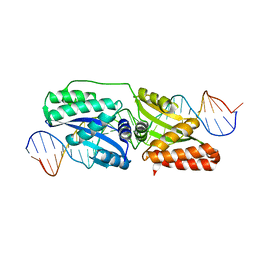 | |
1VS2
 
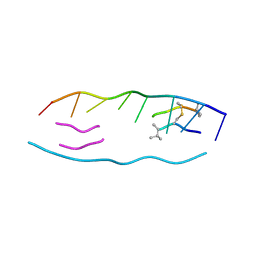 | | Interactions of quinoxaline antibiotic and DNA: the molecular structure of a TRIOSTIN A-D(GCGTACGC) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3', TRIOSTIN A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Rich, A. | | Deposit date: | 1986-10-21 | | Release date: | 2006-06-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of Quinoxaline Antibiotic and DNA: The Molecular Structure of a Triostin A-D(Gcgtacgc) Complex.
J.Biomol.Struct.Dyn., 4, 1986
|
|
1WIJ
 
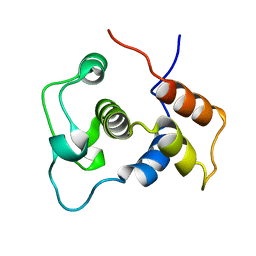 | | Solution Structure of the DNA-Binding Domain of Ethylene-Insensitive3-Like3 | | Descriptor: | ETHYLENE-INSENSITIVE3-like 3 protein | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3.
J.Mol.Biol., 348, 2005
|
|
