8OY0
 
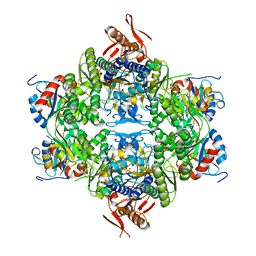 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Acinetobacter baumanii | | Descriptor: | ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, GLYCEROL, ... | | Authors: | Alphey, M.S, Read, B, da Silva, R.G. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.
Biochemistry, 63, 2024
|
|
1SKU
 
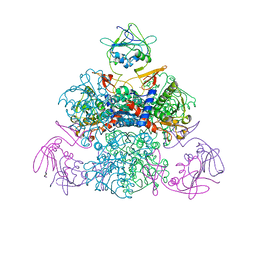 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
4EZU
 
 | |
2BY7
 
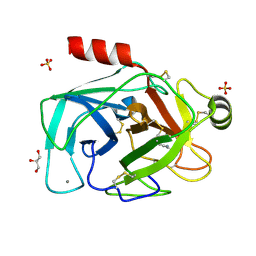 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BYA
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY6
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY9
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY5
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY8
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BLV
 
 | | Trypsin before a high dose x-ray "burn" | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BLW
 
 | | Trypsin after a high dose X-ray "Burn" | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2QCL
 
 | |
2QCM
 
 | |
2QCH
 
 | |
4DNQ
 
 | | Crystal Structure of DAD2 S96A mutant | | Descriptor: | DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DAD2 Is an alpha/beta Hydrolase likely to Be Involved in the Perception of the Plant Branching Hormone, Strigolactone
Curr.Biol., 22, 2012
|
|
2QCN
 
 | |
2XT0
 
 | | Dehalogenase DPpA from Plesiocystis pacifica SIR-I | | Descriptor: | HALOALKANE DEHALOGENASE, SULFATE ION | | Authors: | Bogdanovic, X, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-10-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cloning, Functional Expression, Biochemical Characterization, and Structural Analysis of a Haloalkane Dehalogenase from Plesiocystis Pacifica Sir-1.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
4DNP
 
 | | Crystal Structure of DAD2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, DAD2, GLYCEROL | | Authors: | Hamiaux, C. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DAD2 Is an alpha/beta Hydrolase likely to Be Involved in the Perception of the Plant Branching Hormone, Strigolactone
Curr.Biol., 22, 2012
|
|
2QCD
 
 | |
2QCC
 
 | |
1YHM
 
 | | Structure of the complex of Trypanosoma cruzi farnesyl disphosphate synthase with alendronate, Isopentenyl diphosphate and mg+2 | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
6PRD
 
 | | S. aureus dihydrofolate reductase co-crystallized with para-methoxyphenyl-dimethyoxydihydropthalazine inhibitor and NADP(H) | | Descriptor: | 3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-6,7-dimethoxy-1-(4-methoxyphenyl)-3,4-dihydrophthalazin-2(1H)-yl]propan-1-one, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Thomas, L.M. | | Deposit date: | 2019-07-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inhibitor design to target a unique feature in the folate pocket of Staphylococcus aureus dihydrofolate reductase.
Eur.J.Med.Chem., 200, 2020
|
|
8V0A
 
 | | Crystal Structure of the worst case of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
3PR1
 
 | |
2YBS
 
 | |
