6U7T
 
 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | O'Shea Murray, V.L, Cao, S, Horvath, M.P, David, S.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
1KLO
 
 | |
1KHB
 
 | | PEPCK complex with nonhydrolyzable GTP analog, native data | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KHN
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
4X7E
 
 | |
1KJQ
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-ADP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KKA
 
 | |
1KKK
 
 | |
1KLI
 
 | | Cofactor-and substrate-assisted activation of factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Sichler, K, Banner, D.W, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of Uninhibited Factor VIIa Link its Cofactor and Substrate-assisted Activation to Specific Interactions
J.Mol.Biol., 322, 2002
|
|
8DYD
 
 | | Crystal structure of human SDHA-SDHAF2-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
1KI6
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
5HRU
 
 | |
8DYE
 
 | | Crystal structure of human SDHA-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
1KLV
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor associated protein | | Descriptor: | GABA(A) Receptor associated protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-13 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KNC
 
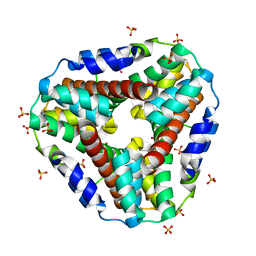 | | Structure of AhpD from Mycobacterium tuberculosis, a novel enzyme with thioredoxin-like activity. | | Descriptor: | AhpD protein, SULFATE ION | | Authors: | Bryk, R, Lima, C.D, Erdjument-Bromage, H, Tempst, P, Nathan, C. | | Deposit date: | 2001-12-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein.
Science, 295, 2002
|
|
5HV4
 
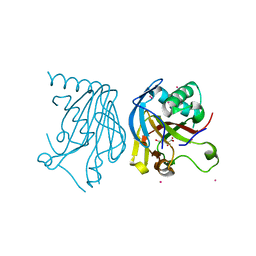 | |
1KJJ
 
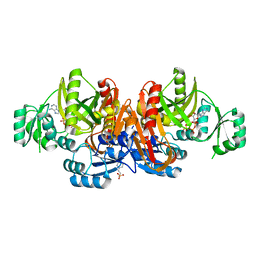 | | Crystal structure of glycniamide ribonucleotide transformylase in complex with Mg-ATP-gamma-S | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KK5
 
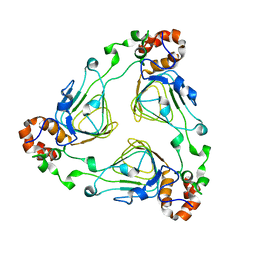 | | Crystal Structure of Vat(D) (Form II) | | Descriptor: | STREPTOGRAMIN A ACETYLTRANSFERASE | | Authors: | Sugantino, M, Roderick, S.L. | | Deposit date: | 2001-12-06 | | Release date: | 2002-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Vat(D): an acetyltransferase that inactivates streptogramin group A antibiotics.
Biochemistry, 41, 2002
|
|
1KKU
 
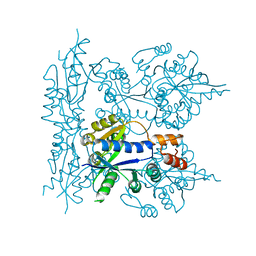 | | Crystal structure of nuclear human nicotinamide mononucleotide adenylyltransferase | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE | | Authors: | Garavaglia, S, D'Angelo, I, Emanuelli, M, Carnevali, F, Pierella, F, Magni, G, Rizzi, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human NMN adenylyltransferase. A key nuclear enzyme for NAD homeostasis.
J.Biol.Chem., 277, 2002
|
|
1KNU
 
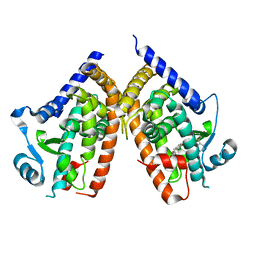 | | LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA IN COMPLEX WITH A SYNTHETIC AGONIST | | Descriptor: | (S)-3-(4-(2-CARBAZOL-9-YL-ETHOXY)-PHENYL)-2-ETHOXY-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Svensson, L.A, Mortensen, S.B, Fleckner, J, Woeldike, H.F. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclic-alpha-alkyloxyphenylpropionic acids: dual PPARalpha/gamma agonists with hypolipidemic and antidiabetic activity
J.MED.CHEM., 45, 2002
|
|
1KO4
 
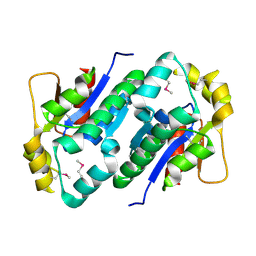 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOK
 
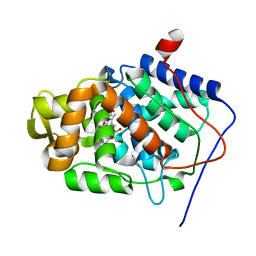 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (MpCcP) | | Descriptor: | Cytochrome c Peroxidase, FE(III)-(4-MESOPORPHYRINONE) | | Authors: | Bhaskar, B, Immoos, C.E, Cohen, M.S, Barrows, T.P, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2001-12-20 | | Release date: | 2002-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mesopone cytochrome c peroxidase: functional model of heme oxygenated oxidases.
J.Inorg.Biochem., 91, 2002
|
|
1KP0
 
 | |
