4GK0
 
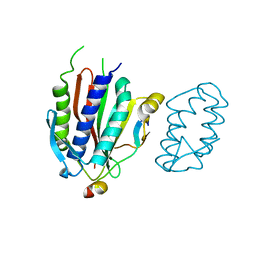 | | Crystal structure of human Rev3-Rev7-Rev1 complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
7Q9U
 
 | |
4FWS
 
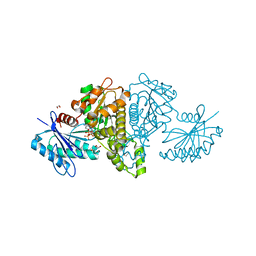 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with CTP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-TRIPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
4GK5
 
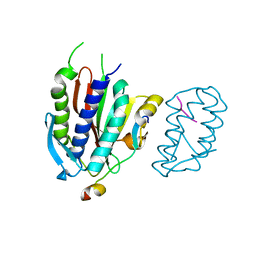 | | Crystal structure of human Rev3-Rev7-Rev1-Polkappa complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
7ZCW
 
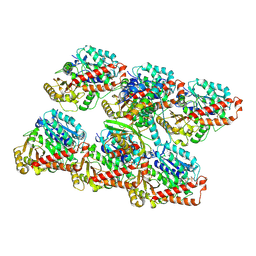 | | Cryo-EM structure of GMPCPP-microtubules in complex with VASH2-SVBP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Choi, S.R, Blum, T, Steinmetz, M.O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | VASH1-SVBP and VASH2-SVBP generate different detyrosination profiles on microtubules.
J.Cell Biol., 222, 2023
|
|
6THA
 
 | | Crystal structure of human sugar transporter GLUT1 (SLC2A1) in the inward conformation | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Solute carrier family 2, ... | | Authors: | Pedersen, B.P, Paulsen, P.A, Custodio, T.F. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of GLUT1 to GLUT3 reveal transport regulation mechanism in sugar porter family.
Life Sci Alliance, 4, 2021
|
|
4H0W
 
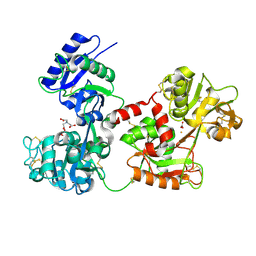 | | Bismuth bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bismuth(III) ION, CARBONATE ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2012-09-10 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe.
Sci Rep, 2, 2012
|
|
8ILL
 
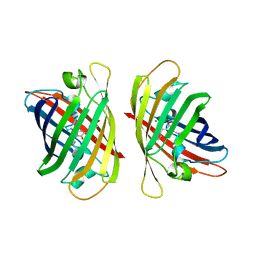 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
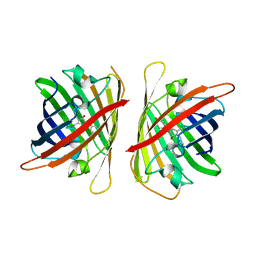 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
7QPD
 
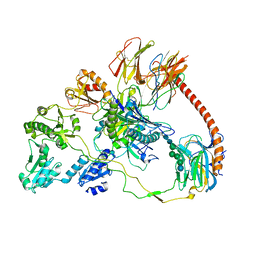 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
6HXU
 
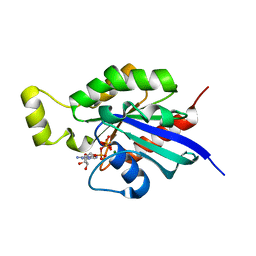 | | Crystal structure of Human RHOB Q63L in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho-related GTP-binding protein RhoB | | Authors: | Soulie, S, Gence, R, Cabantous, S, Lajoie-Mazenc, I, Favre, G, Pedelacq, J.D. | | Deposit date: | 2018-10-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation.
Cell Chem Biol, 26, 2019
|
|
8IVZ
 
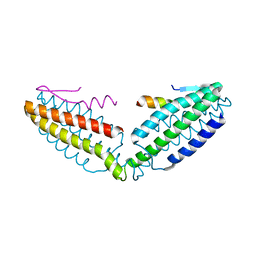 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
4Q6F
 
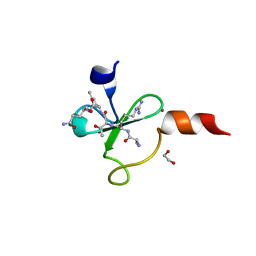 | | Crystal structure of human BAZ2A PHD zinc finger in complex with unmodified H3K4 histone peptide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, ZINC ION, ... | | Authors: | Tallant, C, Overvoorde, L, Krojer, T, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ciulli, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
8IWW
 
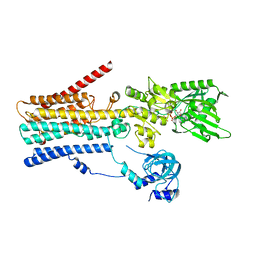 | | hSPCA1 in the CaE1P-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calcium-transporting ATPase type 2C member 1, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWR
 
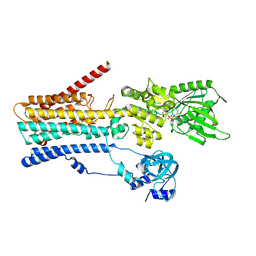 | | hSPCA1 in the CaE1-ATP state | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWP
 
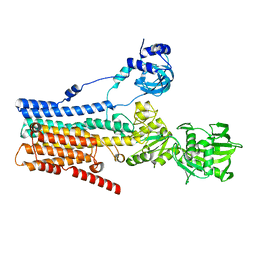 | | hSPCA1 in the CaE1 state | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1 | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWU
 
 | | hSPCA1 in the E2~P state | | Descriptor: | Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWS
 
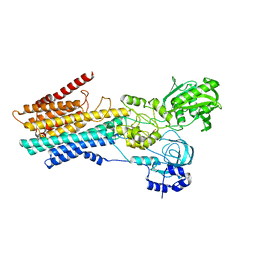 | | hSPCA1 in the CaE2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, Calcium-transporting ATPase type 2C member 1, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8SSU
 
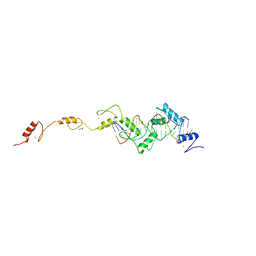 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SST
 
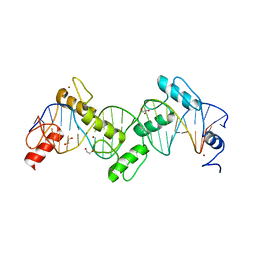 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) K365T Mutant Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
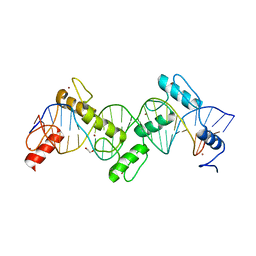 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSQ
 
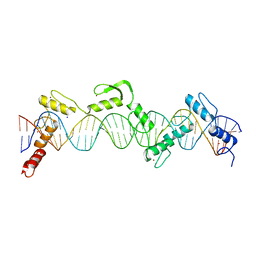 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | Descriptor: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
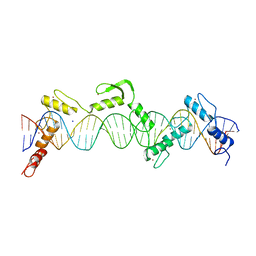 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | Descriptor: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8FO2
 
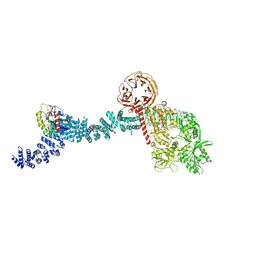 | |
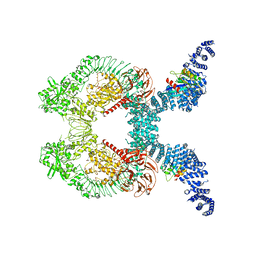
8FO8
 
 | |
