1EGY
 
 | |
4A83
 
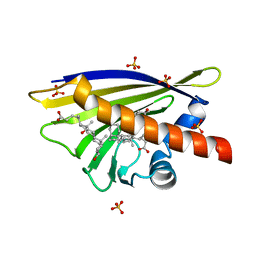 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with deoxycholate. | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
1GUT
 
 | |
2WZK
 
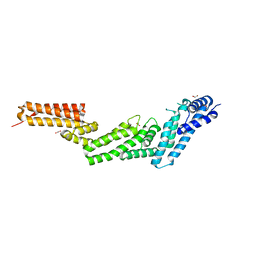 | | Structure of the Cul5 N-terminal domain at 2.05A resolution. | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-5 | | Authors: | Muniz, J.R.C, Ayinampudi, V, Zhang, Y, Babon, J.J, Chaikuad, A, Krojer, T, Pike, A.C.W, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A.N. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
3SG0
 
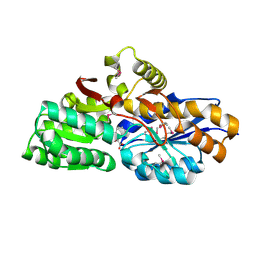 | | The crystal structure of an extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | BENZOYL-FORMIC ACID, Extracellular ligand-binding receptor | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
1F5O
 
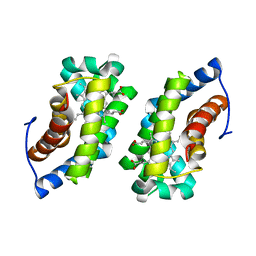 | |
1F28
 
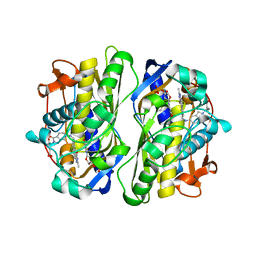 | | CRYSTAL STRUCTURE OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII BOUND TO DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Anderson, A.C, O'Neil, R.H, Surti, T.S, Stroud, R.M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking.
Chem.Biol., 8, 2001
|
|
3BKR
 
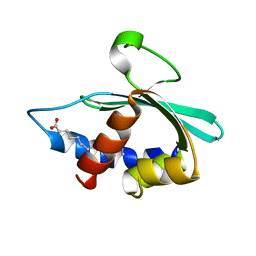 | |
1F5P
 
 | |
1GUO
 
 | |
1GUS
 
 | |
2Q60
 
 | | Crystal structure of the ligand binding domain of polyandrocarpa misakiensis rxr in tetramer in absence of ligand | | Descriptor: | Retinoid X receptor | | Authors: | Borel, F, De Groot, A, Juillan-Binard, C, De Rosny, E, Laudet, V, Pebay-Peyroula, E, Fontecilla-Camps, J.-C, Ferrer, J.-L. | | Deposit date: | 2007-06-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-binding domain of the retinoid X receptor from the ascidian polyandrocarpa misakiensis.
Proteins, 74, 2008
|
|
4XCN
 
 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA); 2.9 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
3KYR
 
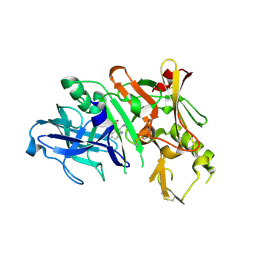 | | Bace-1 in complex with a norstatine type inhibitor | | Descriptor: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | Authors: | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | Deposit date: | 2009-12-07 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
5B41
 
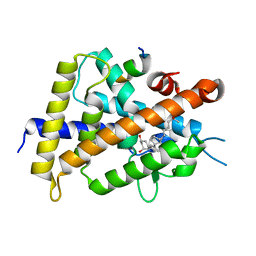 | | Crystal structure of VDR-LBD complexed with 2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Apo- and Antagonist-Binding Structures of Vitamin D Receptor Ligand-Binding Domain Revealed by Hybrid Approach Combining Small-Angle X-ray Scattering and Molecular Dynamics
J.Med.Chem., 59, 2016
|
|
6IAL
 
 | | Porcine E.coli heat-labile enterotoxin B-pentamer in complex with Lacto-N-neohexaose | | Descriptor: | CALCIUM ION, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Heim, J.B, Heggelund, J.E, Krengel, U. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Specificity ofEscherichia coliHeat-Labile Enterotoxin Investigated by Single-Site Mutagenesis and Crystallography.
Int J Mol Sci, 20, 2019
|
|
4XBG
 
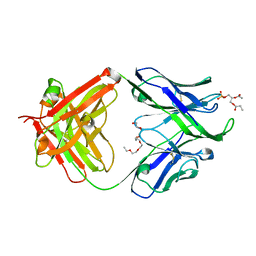 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA): 2.73 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-16 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | Descriptor: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
6ISA
 
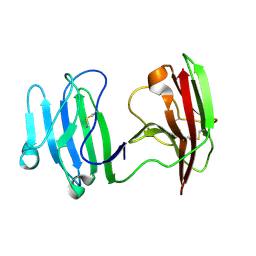 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8EBM
 
 | |
8EBL
 
 | |
4XAW
 
 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41: Crystals cryoprotected with phosphatidic acid (08:0 PA) | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, FRAGMENT OF HIV GLYCOPROTEIN (GP41) including the MPER region 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-15 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
6ISB
 
 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4XC3
 
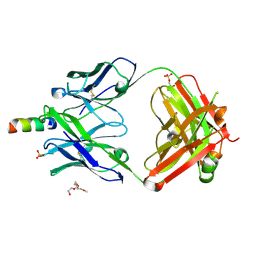 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41; crystals cryoprotected with rac-glycerol 1-phosphate | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, MODIFIED FRAGMENT OF HIV-1 GLYCOPROTEIN (GP41) INCLUDING THE MPER REGION 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XCY
 
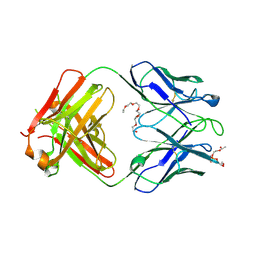 | | Crystal structure of human 4E10 Fab in complex with phosphatidylglycerol (06:0 PG) | | Descriptor: | (2S)-3-{[(R)-{[(2R)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
