9FXD
 
 | |
9FW6
 
 | | A ternary complex of plant adenosine kinase 1 from moss Physcomitrella patens (PpADK1) with adenosine and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kopecny, D, Vigouroux, A, Morera, S. | | Deposit date: | 2024-06-28 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A monomer-dimer switch modulates the activity of plant adenosine kinase.
J.Exp.Bot., 2025
|
|
8W7X
 
 | | SPS_Carbonic Anhydrases | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, SPS_Carbon Anhydrase, ... | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | structure of SPS_Carbon Anhydrase
To Be Published
|
|
8P5R
 
 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
9GIL
 
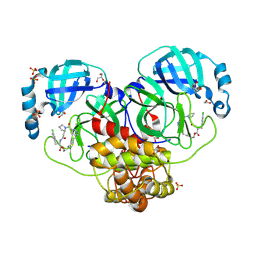 | | Crystal structure of SARS-CoV-2 Mpro with compound 12 | | Descriptor: | (7~{R},11~{R},19~{E})-11-[(4-chlorophenyl)methyl]-13-oxa-3,10,23-triazatricyclo[19.3.1.0^{3,7}]pentacosa-1(24),19,21(25),22-tetraene-2,9,12-trione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Schmitt, A, Preuss, F, Prasad, A, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 67, 2024
|
|
4U53
 
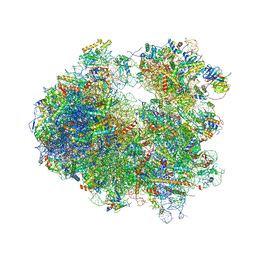 | | Crystal structure of Deoxynivalenol bound to the yeast 80S ribosome | | Descriptor: | (3beta,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U88
 
 | |
4U8H
 
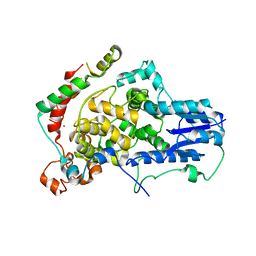 | | Crystal Structure of Mammalian Period-Cryptochrome Complex | | Descriptor: | Cryptochrome-2, Period circadian protein homolog 2, ZINC ION | | Authors: | Nangle, S.N, Rosensweig, C, Koike, N, Tei, H, Takahashi, J.S, Green, C.B, Zheng, N. | | Deposit date: | 2014-08-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex.
Elife, 3, 2014
|
|
4UA9
 
 | |
4UFC
 
 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
4U2M
 
 | |
4U2R
 
 | | Crystal structure of the GLUR2 ligand binding core (S1S2J, flip variant) in the apo state | | Descriptor: | Glutamate receptor 2, SULFATE ION | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4114 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U8U
 
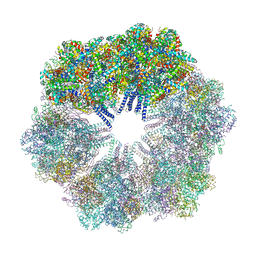 | | The Crystallographic structure of the giant hemoglobin from Glossoscolex paulistus at 3.2 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Bachega, J.F.R, Maluf, F.V, Andi, B, D'Muniz Pereira, H, Carazzollea, M.F, Orville, A, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-08-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U2Y
 
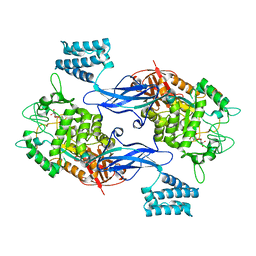 | | Sco GlgEI-V279S in Complex with Reaction Intermediate Azasugar | | Descriptor: | (2R,3R,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)pyrrolidin-3-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1 | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U31
 
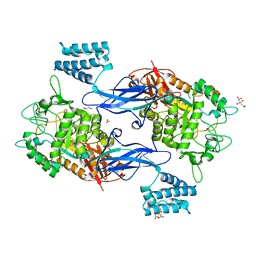 | | Sco GlgEI-V279S in Complex with maltose-C-phosphonate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U3E
 
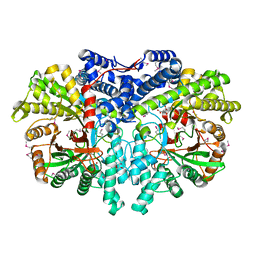 | | Anaerobic ribonucleotide reductase | | Descriptor: | ACETATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-07-20 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The class III ribonucleotide reductase from Neisseria bacilliformis can utilize thioredoxin as a reductant.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4UA4
 
 | |
4UAH
 
 | |
4UB0
 
 | |
4U3P
 
 | | Octameric RNA duplex co-crystallized with strontium(II)chloride | | Descriptor: | RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3'), STRONTIUM ION | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3R
 
 | | Octameric RNA duplex co-crystallized with cobalt(II)chloride | | Descriptor: | COBALT (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3X
 
 | | Structure of a human VH antibody domain binding to the cleft of hen egg lysozyme | | Descriptor: | 1,2-ETHANEDIOL, Human VH domain antibody, Lysozyme C | | Authors: | Rouet, R, Langley, D.B, Christ, D. | | Deposit date: | 2014-07-23 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fully Human VH Single Domains That Rival the Stability and Cleft Recognition of Camelid Antibodies
J.Biol.Chem., 290, 2015
|
|
4UBH
 
 | | Resting state of rat cysteine dioxygenase Y157F variant | | Descriptor: | CHLORIDE ION, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Tchesnokov, E.P, Fellner, M, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of cysteine dioxygenase mutant
To be published
|
|
4UBO
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 3.70 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
