2RRQ
 
 | | DNA oligomer containing propylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*(JDT)P*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
7XKM
 
 | |
4GRE
 
 | |
4GS2
 
 | |
4E2I
 
 | | The Complex Structure of the SV40 Helicase Large T Antigen and p68 Subunit of DNA Polymerase Alpha-Primase | | Descriptor: | DNA polymerase alpha subunit B, Large T antigen, ZINC ION | | Authors: | Zhou, B, Arnett, D.R, Yu, X, Brewster, A, Sowd, G.A, Xie, C.L, Vila, S, Gai, D, Fanning, E, Chen, X.S. | | Deposit date: | 2012-03-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase.
J.Biol.Chem., 287, 2012
|
|
6DBL
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBQ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBV
 
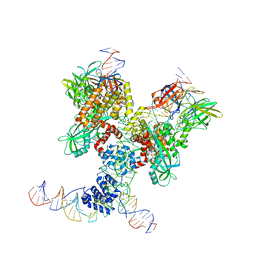 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7WAZ
 
 | |
6AQX
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z4B | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6AQV
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z3B | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
441D
 
 | |
6DBT
 
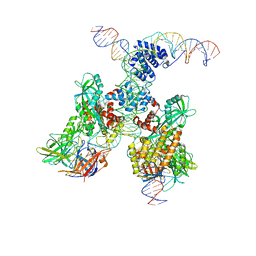 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6EN1
 
 | |
440D
 
 | |
6AQT
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z3A | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6AQW
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z4A | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6EN2
 
 | |
6BWY
 
 | | DNA substrate selection by APOBEC3G | | Descriptor: | DNA (30-MER), PHOSPHATE ION, Protection of telomeres protein 1, ... | | Authors: | Ziegler, S.J, Buzovetsky, O. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into DNA substrate selection by APOBEC3G from structural, biochemical, and functional studies.
PLoS ONE, 13, 2018
|
|
6DBU
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DGD
 
 | | PriA helicase bound to dsDNA of a DNA replication fork | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*CP*CP*GP*AP*CP*T)-3'), DNA (5'-D(P*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*AP*CP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Satyshur, K.A, Windgassen, T.A, Keck, J.L. | | Deposit date: | 2018-05-17 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.823 Å) | | Cite: | Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7WB0
 
 | |
6DBO
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7YGO
 
 | | DNA duplex containing 5OHU-Hg(II)-T base pairs | | Descriptor: | DNA (5'-D(*GP*GP*AP*CP*CP*TP*(6HU)P*GP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Kondo, J, Torigoe, H, Arakawa, F. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Specific binding of Hg 2+ to mismatched base pairs involving 5-hydroxyuracil in duplex DNA.
J.Inorg.Biochem., 241, 2023
|
|
7WAY
 
 | |
