7SPM
 
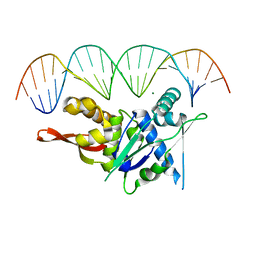 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (30 mg/mL EDC, 12 hours, 2 doses). | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
4XNJ
 
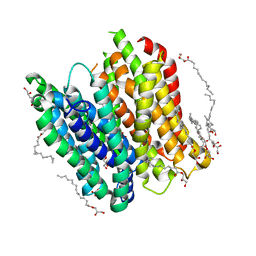 | | X-ray structure of PepTst2 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7CA1
 
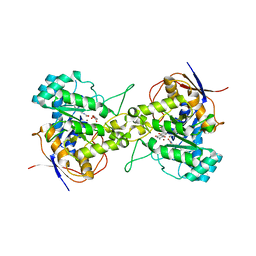 | | Crystal structure of dihydroorotase in complex with plumbagin from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 5-hydroxy-2-methylnaphthalene-1,4-dione, Dihydroorotase, ... | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plumbagin, a Natural Product with Potent Anticancer Activities, Binds to and Inhibits Dihydroorotase, a Key Enzyme in Pyrimidine Biosynthesis.
Int J Mol Sci, 22, 2021
|
|
7CA0
 
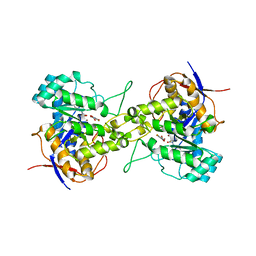 | | Crystal structure of dihydroorotase in complex with 5-fluoroorotic acid from Saccharomyces cerevisiae | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complexed Crystal Structure of Saccharomyces cerevisiae Dihydroorotase with Inhibitor 5-Fluoroorotate Reveals a New Binding Mode.
Bioinorg Chem Appl, 2021, 2021
|
|
6UCK
 
 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Bent Conformer | | Descriptor: | Islet amyloid polypeptide | | Authors: | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | Deposit date: | 2019-09-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6UCJ
 
 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Open Conformer | | Descriptor: | Islet amyloid polypeptide | | Authors: | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | Deposit date: | 2019-09-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
7CET
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEU
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
6MVF
 
 | |
8Z4B
 
 | |
6S5V
 
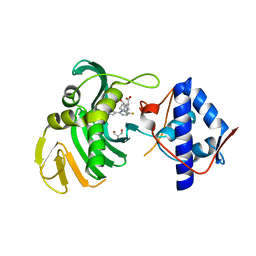 | | Crystal structure of the Cap-Midlink region of the H5N1 Influenza A virus polymerase in complex with a Cap-domain binding analogue | | Descriptor: | (1~{S},2~{S},3~{S},6~{R})-2-[[2-[5,7-bis(fluoranyl)-1~{H}-indol-3-yl]-5-fluoranyl-pyrimidin-4-yl]amino]-3,6-dimethyl-cyclohexane-1-carboxylic acid, GLYCEROL, POTASSIUM ION, ... | | Authors: | Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-07-02 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Novel Indoles Targeting the Influenza PB2 Cap Binding Region.
J.Med.Chem., 62, 2019
|
|
12E8
 
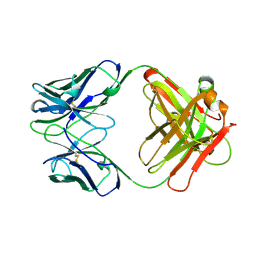 | | 2E8 FAB FRAGMENT | | Descriptor: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | Authors: | Rupp, B, Trakhanov, S. | | Deposit date: | 1998-03-14 | | Release date: | 1998-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
6MVG
 
 | |
4XNI
 
 | | X-ray structure of PepTst1 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
6UX5
 
 | |
9GNS
 
 | | X-ray structure of Human holo aromatic L-amino acid decarboxylase (AADC) complex with Carbidopa at physiological pH | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CARBIDOPA, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Perduca, M, Bisello, G, Bertoldi, M. | | Deposit date: | 2024-09-04 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
5LIT
 
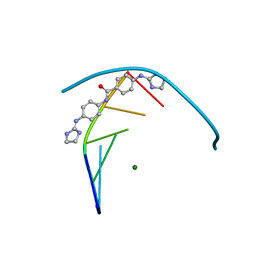 | | Structure of the DNA duplex d(AAATTT)2 with the potential antiparasitic drug 6XV at 1.25 A resolution | | Descriptor: | 4-((4,5-dihydro-1H-imidazol-2-yl)amino)-N-(4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)benzamide dihydrochloride, DNA (5'-D(*AP*AP*AP*TP*TP*T)-3'), MAGNESIUM ION | | Authors: | Millan, C.R, Dardonville, C, de Koning, H.P, Saperas, N, Lourdes Campos, J. | | Deposit date: | 2016-07-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res., 45, 2017
|
|
1AM5
 
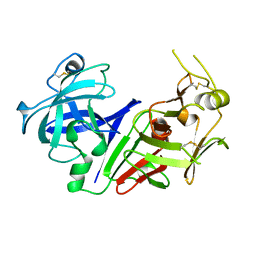 | |
6NEE
 
 | |
9BYJ
 
 | | Crystal Structure of Hck in complex with the Src-family kinase inhibitor A-419259 | | Descriptor: | 1,2-ETHANEDIOL, 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Selzer, A.M, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2024-05-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystallization of the Src-Family Kinase Hck with the ATP-Site Inhibitor A-419259 Stabilizes an Extended Activation Loop Conformation.
Biochemistry, 63, 2024
|
|
9CQE
 
 | |
3ES7
 
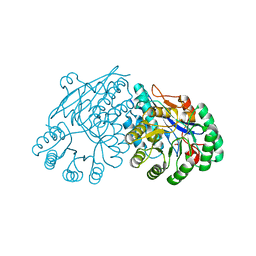 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3ES8
 
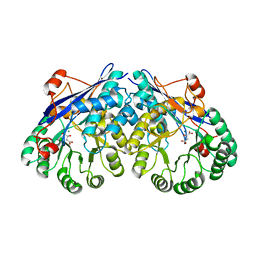 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
