5I2G
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | Diol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
4CM4
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5-(4-fluorophenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
5I3U
 
 | |
4WZ5
 
 | | Crystal structure of P. aeruginosa OXA10 | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
Acs Infect Dis., 1, 2015
|
|
4WYY
 
 | | Crystal Structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
5I9Q
 
 | | Crystal structure of 3BNC55 Fab in complex with 426c.TM4deltaV1-3 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC55 Fab heavy chain, ... | | Authors: | Scharf, L, Chen, C, Bjorkman, P.J. | | Deposit date: | 2016-02-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
4UAU
 
 | | Crystal structure of CbbY (mutant D10N) from Rhodobacter sphaeroides in complex with Xylulose-(1,5)bisphosphate, crystal form II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Protein CbbY, ... | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
9EYU
 
 | | Human PRMT5 in complex with AZ compound 1 | | Descriptor: | (1~{S})-2-[(2-carbamimidamido-1,3-thiazol-5-yl)methyl]-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
4WRH
 
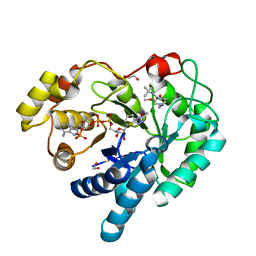 | | AKR1C3 complexed with breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(aminomethyl)-2-chloro-6-methoxyphenoxy]-N-tert-butylacetamide, 5-methyl-4H-1,2,4-triazole-3-thiol, ... | | Authors: | Squire, C.J, Flanagan, J.U, Yosaatmadja, Y. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide trapped in active site of AKR1C3
To Be Published
|
|
6UTD
 
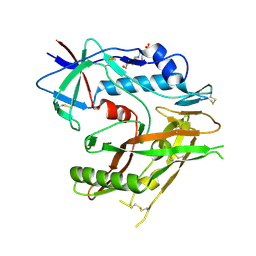 | |
4UMX
 
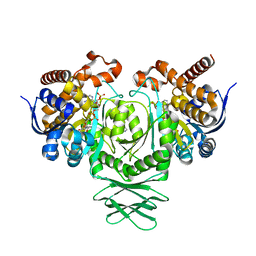 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | 2,6-bis(1H-imidazol-1-ylmethyl)-4-(2,4,4-trimethylpentan-2-yl)phenol, GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, ... | | Authors: | Mathieu, M, Marquette, J.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
6UTB
 
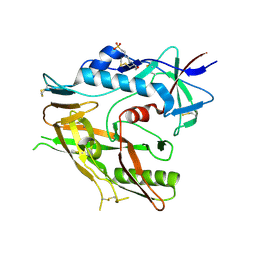 | |
6USW
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210 | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
4UEK
 
 | | Galactitol-1-phosphate 5-dehydrogenase from E. coli with Tris within the active site. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UEJ
 
 | | Closed state of galactitol-1-phosphate 5-dehydrogenase from E. coli in complex with glycerol. | | Descriptor: | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, GLYCEROL, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
7STZ
 
 | |
9IZL
 
 | | hVanin-1 complexed with X17 | | Descriptor: | 8-oxa-2-azaspiro[4.5]decan-2-yl-[2-[(3~{S})-3-oxidanylpyrrolidin-1-yl]-1,3-thiazol-5-yl]methanone, Pantetheinase | | Authors: | Fan, S, Zhen, L, Xie, T. | | Deposit date: | 2024-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Thiazole Carboxamides as Novel Vanin-1 Inhibitors for Inflammatory Bowel Disease Treatment.
J.Med.Chem., 67, 2024
|
|
4WZ4
 
 | | Crystal structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, GLYCEROL, {(3R)-6-[(3-amino-1,2,4-thiadiazol-5-yl)oxy]-1-hydroxy-4,5-dimethyl-1,3-dihydro-2,1-benzoxaborol-3-yl}acetic acid | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
1EU1
 
 | | THE CRYSTAL STRUCTURE OF RHODOBACTER SPHAEROIDES DIMETHYLSULFOXIDE REDUCTASE REVEALS TWO DISTINCT MOLYBDENUM COORDINATION ENVIRONMENTS. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Li, H.K, Temple, K, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-04-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of Rhodobacter sphaeroides Dimethylsulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments
J.Am.Chem.Soc., 122, 2000
|
|
7PXX
 
 | | The crystal structure of Leishmania major Pteridine Reductase 1 in complex with substrate folic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, FOLIC ACID, GLYCEROL, ... | | Authors: | Di Pisa, F, Dello Iacono, L, Mangani, S. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the ternary complex of Leishmania major pteridine reductase 1 with the cofactor NADP + /NADPH and the substrate folic acid.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8GJT
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with TFH-I-116-D1 | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-[(3R,5S)-3,4,5-trimethylpiperazine-1-carbonyl]piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GCZ
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with ZXC-I-090 | | Descriptor: | (3S,5S)-5-(aminomethyl)-N-(4-chloro-3-fluorophenyl)-1-(4-methylpiperazine-1-carbonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nguyen, D.N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD5
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD3
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-101 | | Descriptor: | (3S,5R)-N-(4-chloro-3-fluorophenyl)-1-(4-glycylpiperazine-1-carbonyl)-5-(hydroxymethyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
