2QKK
 
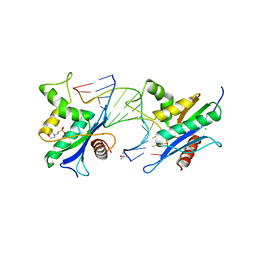 | | Human RNase H catalytic domain mutant D210N in complex with 14-mer RNA/DNA hybrid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3', ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-11 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2NQH
 
 | |
384D
 
 | |
8TAC
 
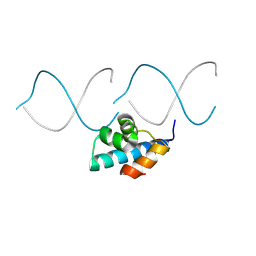 | | Designed DNA binding protein | | Descriptor: | DBP48, DNA (5'-D(*AP*CP*CP*TP*GP*AP*CP*GP*CP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*GP*TP*CP*AP*GP*GP)-3') | | Authors: | Doyle, L, Stoddard, B.L, Glasscock, C.J, McHugh, R.P, Pecoraro, R.J. | | Deposit date: | 2023-06-27 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Designed DNA binding protein
To Be Published
|
|
2BQ2
 
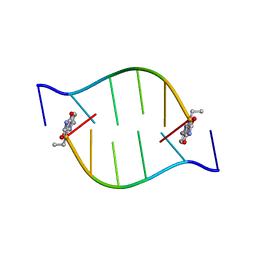 | | Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Linked Nalidixic Acid Residue at the 3' Terminal Nucleotide | | Descriptor: | 5'-D(*AP*CP*GP*CP*GP*2AU)-3', NALIDIXIC ACID | | Authors: | Siegmund, K, Maheshwary, S, Narayanan, S, Connors, W, Richert, M. | | Deposit date: | 2005-04-26 | | Release date: | 2006-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it.
Nucleic Acids Res., 33, 2005
|
|
6YXQ
 
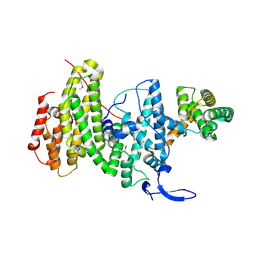 | | Crystal structure of a DNA repair complex ASCC3-ASCC2 | | Descriptor: | Activating signal cointegrator 1 complex subunit 2, Activating signal cointegrator 1 complex subunit 3 | | Authors: | Jia, J, Absmeier, E, Holton, N, Bohnsack, K.E, Pietrzyk-Brzezinska, A.J, Bohnsack, M.T, Wahl, M.C. | | Deposit date: | 2020-05-03 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.
Nat Commun, 11, 2020
|
|
1YNW
 
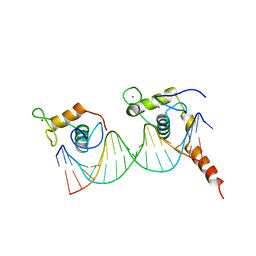 | |
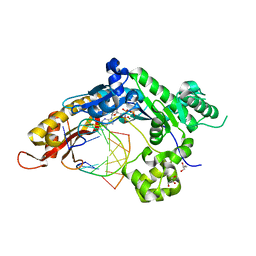
6Q02
 
 | |
6FE8
 
 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | Deposit date: | 2017-12-30 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
7LTN
 
 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | Descriptor: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1PRA
 
 | |
2VHG
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Right End 31-mer DNA | | Descriptor: | MANGANESE (II) ION, RIGHT END 31-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2ODH
 
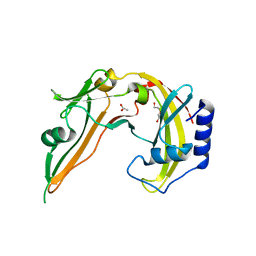 | | Restriction Endonuclease BCNI in the Absence of DNA | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, R.BcnI | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA.
J.Mol.Biol., 369, 2007
|
|
5YTY
 
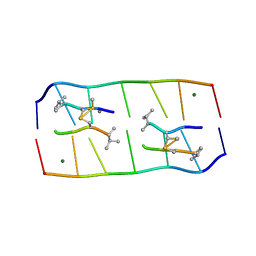 | | Crystal structure of echinomycin-d(ACGACGT/ACGTCGT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*AP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Wu, P.C, Kao, Y.F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Cooperative recognition of T:T mismatch by echinomycin causes structural distortions in DNA duplex
Nucleic Acids Res., 46, 2018
|
|
6CFX
 
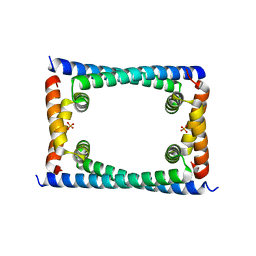 | | Bosea sp GapR solved in the presence of DNA | | Descriptor: | PHOSPHATE ION, UPF0335 protein ASE63_04290 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
2HXM
 
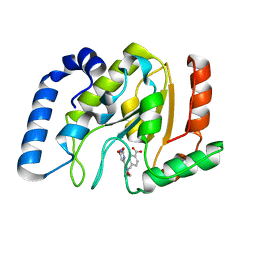 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
2VJU
 
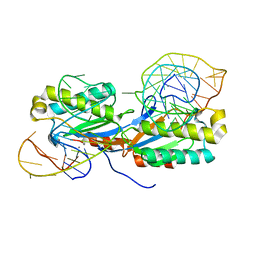 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2IBT
 
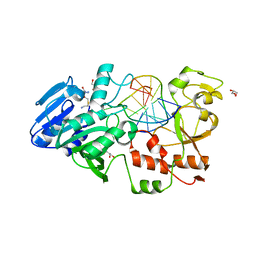 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position and an abasic site analog at the target base partner position | | Descriptor: | 5'-D(*GP*AP*CP*AP*(3DR)P*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
4RNO
 
 | |
3GE4
 
 | |
1LFB
 
 | | THE X-RAY STRUCTURE OF AN ATYPICAL HOMEODOMAIN PRESENT IN THE RAT LIVER TRANSCRIPTION FACTOR LFB1(SLASH)HNF1 AND IMPLICATIONS FOR DNA BINDING | | Descriptor: | LIVER TRANSCRIPTION FACTOR (LFB1) | | Authors: | Ceska, T.A, Lamers, M, Monaci, P, Nicosia, A, Cortese, R, Suck, D. | | Deposit date: | 1993-06-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding.
EMBO J., 12, 1993
|
|
3GDD
 
 | | An inverted anthraquinone-DNA crystal structure | | Descriptor: | 5'-D(*(BRU)P*AP*GP*G)-3', MAGNESIUM ION, N,N'-(9,10-dioxo-9,10-dihydroanthracene-2,7-diyl)bis[2-(dimethylamino)acetamide] | | Authors: | Subirana, J.A, De Luchi, D, Wright, G, Gouyette, C. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a stacked anthraquinone-DNA complex
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2HOS
 
 | | Phage-Selected Homeodomain Bound to Unmodified DNA | | Descriptor: | 3-METHYL-1,3-OXAZOLIDIN-2-ONE, 5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*CP*CP*GP*GP*A)-3', ... | | Authors: | Shokat, K.M, Feldman, M.E, Simon, M.D. | | Deposit date: | 2006-07-16 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and properties of a re-engineered homeodomain protein-DNA interface.
Acs Chem.Biol., 1, 2006
|
|
6AIL
 
 | | CRYSTAL STRUCTURE AT 1.3 ANGSTROMS RESOLUTION OF A NOVEL UDG, UdgX, FROM Mycobacterium smegmatis | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase X | | Authors: | Ahn, W.C, Aroli, S, Varshney, V, Woo, E.J. | | Deposit date: | 2018-08-24 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.335 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
1IU3
 
 | | CRYSTAL STRUCTURE OF THE E.COLI SEQA PROTEIN COMPLEXED WITH HEMIMETHYLATED DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*TP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-26 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein.
Nucleic Acids Res., 32, 2004
|
|
