1MC3
 
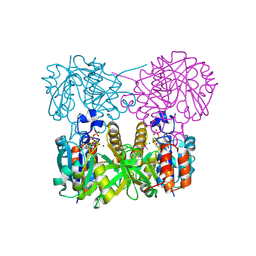 | | CRYSTAL STRUCTURE OF RFFH | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-08-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli Glucose-1-Phosphate Thymidylyltransferase (RffH) Complexed with dTTP and Mg2+
J.BIOL.CHEM., 277, 2002
|
|
9H2D
 
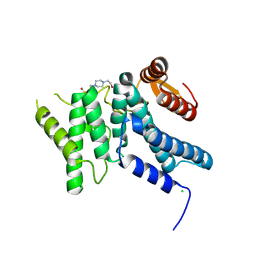 | |
9GUW
 
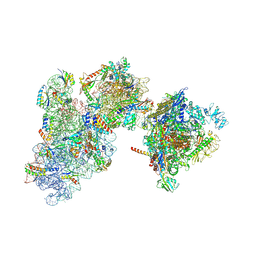 | | 30S-TEC (TEC in expressome position) Inactive state 2 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
1MCH
 
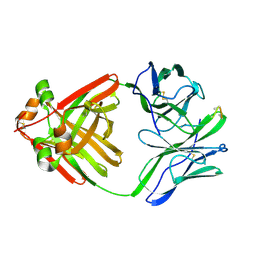 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCQ
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
9H5J
 
 | | PAD12 0N3R tau PHF seeded by AD | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Lovestam, S, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-10-22 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Twelve phosphomimetic mutations induce the assembly of recombinant full-length human tau into paired helical filaments
To Be Published
|
|
1M49
 
 | | Crystal Structure of Human Interleukin-2 Complexed with SP-1985 | | Descriptor: | 2-[2-(1-CARBAMIMIDOYL-PIPERIDIN-3-YL)-ACETYLAMINO]-3-{4-[2-(3-OXALYL-1H-INDOL-7-YL)ETHYL]-PHENYL}-PROPIONIC ACID METHYL ESTER, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
9H5G
 
 | | PAD12 0N3R:0N4R tau PHF | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Lovestam, S, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-10-22 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Twelve phosphomimetic mutations induce the assembly of recombinant full-length human tau into paired helical filaments
To Be Published
|
|
9GUS
 
 | | 30S mRNA delivery complex TEC resolved (30S only) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
9H1P
 
 | |
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MD8
 
 | | Monomeric structure of the active catalytic domain of complement protease C1r | | Descriptor: | C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
9GUR
 
 | | 30S mRNA delivery complex TEC resolved (TEC only) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
9GUQ
 
 | | 30S PIC (Pre-Initiation complex) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
1ME5
 
 | |
9GWO
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571126 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-27 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GUU
 
 | | 30S mRNA delivery complex (consensus) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
9H3E
 
 | | Hen egg white lysozyme crystallization and structure determination at room temperature in the CrystalChip | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Pachl, P, Coudray, L, VIncent, R, Sauter, C. | | Deposit date: | 2024-10-16 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein crystallization and structure determination at room temperature in the CrystalChip.
Febs Open Bio, 15, 2025
|
|
1MF2
 
 | | ANTI HIV1 PROTEASE FAB COMPLEX | | Descriptor: | MONOCLONAL ANTIBODY F11.2.32 | | Authors: | Lescar, J, Bentley, G.A. | | Deposit date: | 1996-12-27 | | Release date: | 1997-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of an Fab-peptide complex: structural basis of HIV-1 protease inhibition by a monoclonal antibody.
J.Mol.Biol., 267, 1997
|
|
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
1ZWJ
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana AT5G18200 | | Descriptor: | ZINC ION, putative galactose-1-phosphate uridyl transferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, McCoy, J.G, Johnson, K.A, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
7S8S
 
 | | Crystal Structure of HLA A*1101 in complex with RVLVNGTFLK, an 10-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
2P4Q
 
 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
7S8R
 
 | | Crystal Structure of HLA A*1101 in complex with SALEWIKNK, an 9-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
