5M6L
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[6-[(4-fluorophenyl)methyl]-3,3-dimethyl-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-[(2~{R},5~{R})-5-methyl-2-[[(3~{R})-3-methylmorpholin-4-yl]methyl]piperazin-4-ium-1-yl]ethanone, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
5M6M
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[[(2~{R},5~{R})-1-[2-[3,3-dimethyl-6-(phenylmethyl)-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-oxidanylidene-ethyl]-5-methyl-piperazin-4-ium-2-yl]methyl]pyrrolidin-2-one, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
5M65
 
 | |
1KNI
 
 | | Stabilizing Disulfide Bridge Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Jacobson, R.H, Matsumura, M, Faber, H.R, Matthews, B.W. | | Deposit date: | 2001-12-18 | | Release date: | 2001-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a stabilizing disulfide bridge mutant that closes the active-site cleft of T4 lysozyme.
Protein Sci., 1, 1992
|
|
4MVA
 
 | | 1.43 Angstrom Resolution Crystal Structure of Triosephosphate Isomerase (tpiA) from Escherichia coli in Complex with Acetyl Phosphate. | | Descriptor: | 1,2-ETHANEDIOL, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M.L, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
8CHP
 
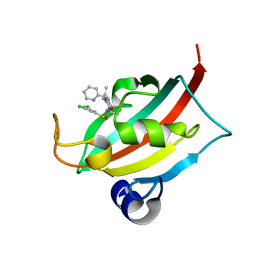 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfinyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfinyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
1KRH
 
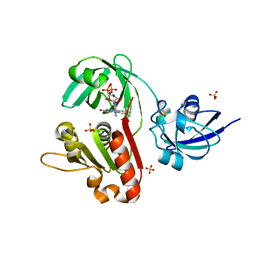 | | X-ray Structure of Benzoate Dioxygenase Reductase | | Descriptor: | Benzoate 1,2-Dioxygenase Reductase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Karlsson, A, Beharry, Z.M, Eby, D.M, Coulter, E.D, Niedle, E.L, Kurtz Jr, D.M, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1.
J.Mol.Biol., 318, 2002
|
|
5FED
 
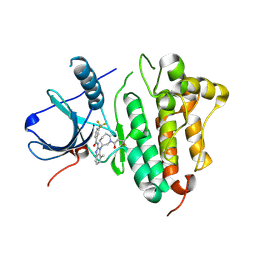 | | EGFR kinase domain in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
5FEE
 
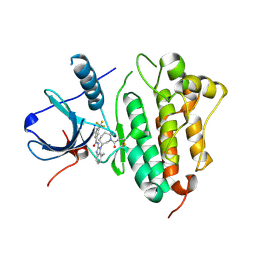 | | EGFR kinase domain T790M mutant in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
5M6E
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[3,3-dimethyl-6-(phenylmethyl)-2~{H}-pyrrolo[3,2-c]pyridin-1-yl]-2-[(2~{R},5~{R})-5-methyl-2-[(4-methylpyrazol-1-yl)methyl]piperazin-4-ium-1-yl]ethanone, DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase XIAP, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
8CNR
 
 | | Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus as isolated in a reduced state at 1.45-A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon.
Front Microbiol, 14, 2023
|
|
6T8Z
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
1KFY
 
 | | QUINOL-FUMARATE REDUCTASE WITH QUINOL INHIBITOR 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL | | Descriptor: | 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-24 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
8CNS
 
 | | The Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus in a mixed redox state after soaking with hydroxylamine, at 1.36-A resolution. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon.
Front Microbiol, 14, 2023
|
|
8QMV
 
 | | L2 forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes.
Embo J., 44, 2025
|
|
6T0H
 
 | | Crystal structure of CYP124 in complex with 1-alpha-hydroxy-vitamin D3 | | Descriptor: | 1-alpha-hydroxy-vitamin D3, CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
8QG5
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-(phenylmethyl)purin-8-yl]prop-2-ynyl-methyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To be published
|
|
8QG6
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[3-[6-azanyl-9-(phenylmethyl)purin-8-yl]prop-2-ynoxymethyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To be published
|
|
6T92
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6DUB
 
 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
5FEC
 
 | |
2YY7
 
 | | Crystal structure of thermolabile L-threonine dehydrogenase from Flavobacterium frigidimaris KUC-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, ... | | Authors: | Yoneda, K, Sakuraba, H, Oikawa, T, Muraoka, I, Ohshima, T. | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Crystal structure of UDP-galactose 4-epimerase-like L-threonine dehydrogenase belonging to the intermediate short-chain dehydrogenase-reductase superfamily
Febs J., 277, 2010
|
|
7A1Q
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
8QMW
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitutions R269W, E271R, L273N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes.
Embo J., 44, 2025
|
|
6DOI
 
 | |
