1K1M
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N(3-AMIDINO-L-PHENYLALANINYL)-4-ACETYL-PIPERAZINE, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1P
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, [((1R)-2-{(2S)-2-[({4-[AMINO(IMINO)METHYL]BENZYL}AMINO)CARBONYL]AZETIDINYL}-1-CYCLOHEXYL-2-OXOETHYL)AMINO]ACETIC ACID | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
6XSW
 
 | | Structure of the Notch3 NRR in complex with an antibody Fab Fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-N3 Fab Heavy Chain, ... | | Authors: | Bard, J. | | Deposit date: | 2020-07-16 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | NOTCH3-targeted antibody drug conjugates regress tumors by inducing apoptosis in receptor cells and through transendocytosis into ligand cells.
Cell Rep Med, 2, 2021
|
|
6DJZ
 
 | | Human sigma-1 receptor bound to haloperidol | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, GLYCEROL, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1DTG
 
 | | HUMAN TRANSFERRIN N-LOBE MUTANT H249E | | Descriptor: | CARBONATE ION, FE (III) ION, TRANSFERRIN | | Authors: | MacGillivray, R.T, Bewley, M.C, Smith, C.A, He, Q.Y, Mason, A.B. | | Deposit date: | 2000-01-12 | | Release date: | 2000-01-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutation of the iron ligand His 249 to Glu in the N-lobe of human transferrin abolishes the dilysine "trigger" but does not significantly affect iron release.
Biochemistry, 39, 2000
|
|
5KWV
 
 | |
8CDO
 
 | |
2J3Q
 
 | | Torpedo acetylcholinesterase complexed with fluorophore thioflavin T | | Descriptor: | 2-[4-(DIMETHYLAMINO)PHENYL]-6-HYDROXY-3-METHYL-1,3-BENZOTHIAZOL-3-IUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Harel, M, Cusack, B, Johnson, J.L, Silman, I, Sussman, J.L, Rosenberry, T.L. | | Deposit date: | 2006-08-23 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site.
J. Am. Chem. Soc., 130, 2008
|
|
8C6Y
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 2 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CB9
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with D-Glucose-2-phosphate | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-25 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
9CB3
 
 | | E2F1-Cyclin F Interface | | Descriptor: | Cyclin-F, E2F1 peptide, S-phase kinase-associated protein 1 | | Authors: | Ngoi, P, Serrao, V.H, Rubin, S.M. | | Deposit date: | 2024-06-18 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural mechanism for the recognition of E2F1 by the ubiquitin ligase adaptor Cyclin F.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6NFJ
 
 | |
9GFE
 
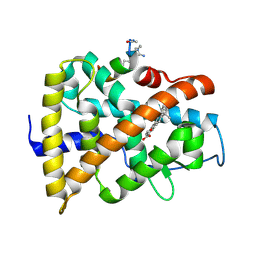 | | hRAR LBD protein in complex with AM580 agonist ligand and a stapled peptide | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, Retinoic acid receptor alpha, Stapled peptide-like ligand | | Authors: | Perdriau, C, Luton, A, Zimmeter, K, Neuville, M, Saragaglia, C, Peluso-lltis, C, Kauffmann, B, Collie, G, Rochel, N, Guichard, G, Pasco, M. | | Deposit date: | 2024-08-09 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Guanidinium-Stapled Helical Peptides for Targeting Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9H9X
 
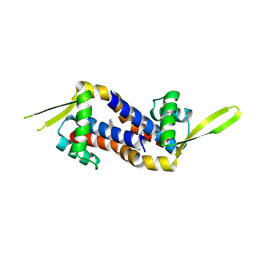 | | Crystal structure of metal-free LmrR_V15Bpy in a closed state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | An Artificial Copper-Michaelase Featuring a Genetically Encoded Bipyridine Ligand for Asymmetric Additions to Nitroalkenes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9H9Z
 
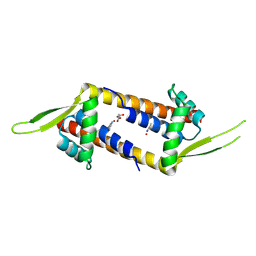 | | Crystal structure of Cu(II)-bound LmrR_V15Bpy | | Descriptor: | COPPER (II) ION, MALONATE ION, Transcriptional regulator, ... | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Artificial Copper-Michaelase Featuring a Genetically Encoded Bipyridine Ligand for Asymmetric Additions to Nitroalkenes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8R41
 
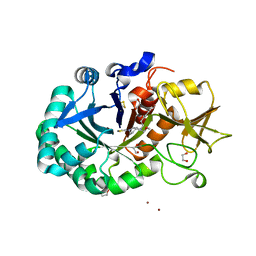 | | Structure of CHI3L1 in complex with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8R4X
 
 | | Structure of Chitinase-3-like protein 1 in complex with inhibitor 30 | | Descriptor: | (2~{S},5~{S})-4-[1-(4-chloranylpyridin-2-yl)piperidin-4-yl]-5-[(4-chlorophenyl)methyl]-2-methyl-morpholine, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8R42
 
 | | Structure of CHI3L1 in complex with inhibititor 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R})-2-[(4-chlorophenyl)methyl]pyrrolidin-1-yl]piperidin-1-yl]pyridine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
6WPE
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SL4
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
3SPS
 
 | |
8A55
 
 | |
5TWX
 
 | | Crystal Structure of BRD9 bromodomain | | Descriptor: | Bromodomain-containing protein 9, N-[6-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)hexyl]-2-(4-{2-[N-(1,1-dioxo-1lambda~6~-thian-4-yl)carbamimidoyl]-5-methyl-4-oxo-4,5-dihydrothieno[3,2-c]pyridin-7-yl}-2-methoxyphenoxy)acetamide | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8CAY
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Siragu, S. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
