6IH5
 
 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
6IH2
 
 | |
6IH3
 
 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH8
 
 | |
6IH6
 
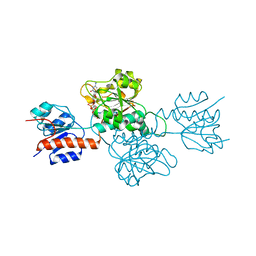 | | Phosphite Dehydrogenase mutant I151R/P176R/M207A from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6ABI
 
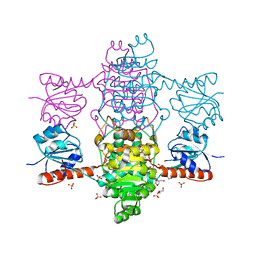 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6ABJ
 
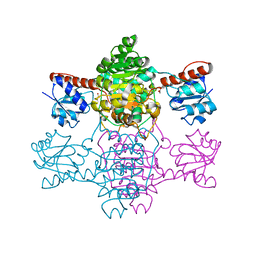 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
5Z20
 
 | | The ternary structure of D-lactate dehydrogenase from Pseudomonas aeruginosa with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase (Fermentative), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
5Z21
 
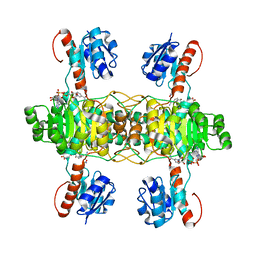 | | The ternary structure of D-lactate dehydrogenase from Fusobacterium nucleatum with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase, OXAMIC ACID | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
5Z1Z
 
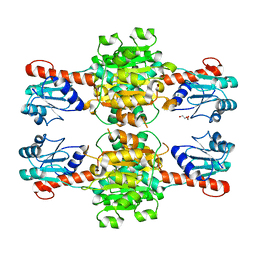 | | The apo-structure of D-lactate dehydrogenase from Escherichia coli | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
6CDF
 
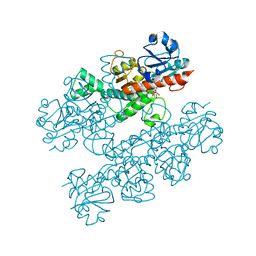 | | Human CtBP1 (28-378) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
6CDR
 
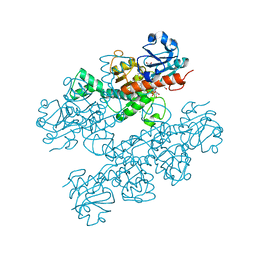 | | Human CtBP1 (28-378) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
6BII
 
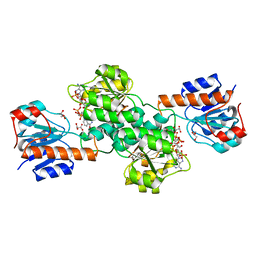 | | Crystal Structure of Pyrococcus yayanosii Glyoxylate Hydroxypyruvate Reductase in complex with NADP and malonate (re-refinement of 5AOW) | | Descriptor: | GLYCEROL, Glyoxylate reductase, MALONATE ION, ... | | Authors: | Lassalle, L, Shabalin, I.G, Girard, E, Minor, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases.
Sci Rep, 6, 2016
|
|
5N6C
 
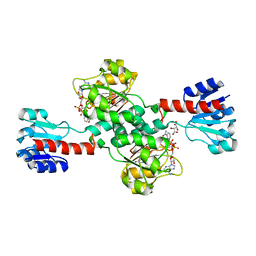 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with NAD and L-Tartrate | | Descriptor: | D-3-phosphoglycerate dehydrogenase, L(+)-TARTARIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Unterlass, J.E, Basle, A, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-02-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the enzymatic activity and potential substrate promiscuity of human 3-phosphoglycerate dehydrogenase (PHGDH).
Oncotarget, 8, 2017
|
|
5OFW
 
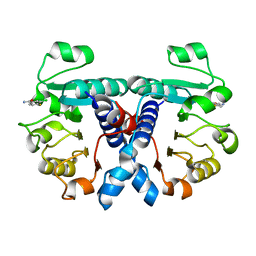 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Chloro-4-fluorobenzamide | | Descriptor: | 3-chloranyl-4-fluoranyl-benzamide, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFV
 
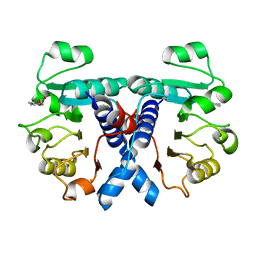 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-fluoro-2-methylbenzoic acid | | Descriptor: | 5-fluoranyl-2-methyl-benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFM
 
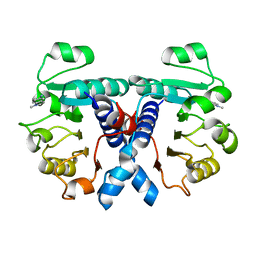 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole | | Descriptor: | 1-methylindol-5-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole
To be published
|
|
5NZQ
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-(1,3-oxazol-5-yl)aniline. | | Descriptor: | 3-(1,3-oxazol-5-yl)aniline, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NZP
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Hydroxybenzisoxazole | | Descriptor: | 1,2-benzoxazol-3-ol, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NZO
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 1-methyl-3-phenyl-1H-pyrazol-5-amine | | Descriptor: | 2-methyl-5-phenyl-pyrazol-3-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5N53
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with N-(3-chloro-4-methoxyphenyl) acetamide | | Descriptor: | D-3-phosphoglycerate dehydrogenase, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-02-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5V7G
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Mason, D.V, Handing, K.B, Kutner, J, Matelska, D, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V7N
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and 2-Keto-D-gluconic acid | | Descriptor: | 2-keto-D-gluconic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miks, C.D, Kutner, J, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V6Q
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and malonate | | Descriptor: | MALONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent glyoxylate/hydroxypyruvate reductase, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miezaniec, A.P, Gasiorowska, O.A, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
