3M9A
 
 | | Protein structure of type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1OL0
 
 | | Crystal structure of a camelised human VH | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN G, SULFATE ION | | Authors: | Dottorini, T, Vaughan, C.K, Walsh, M.A, Losurdo, P, Sollazzo, M. | | Deposit date: | 2003-08-02 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Human Vh: Requirements for Maintaining a Monomeric Fragment
Biochemistry, 43, 2004
|
|
4XH2
 
 | | Crystal structure of human paxillin LD4 motif in complex with Fab fragment | | Descriptor: | ACETATE ION, ACETYL GROUP, Fab Heavy Chain, ... | | Authors: | Nocula-Lugowska, M, Lugowski, M, Salgia, R, Kossiakoff, A.A. | | Deposit date: | 2015-01-04 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering Synthetic Antibody Inhibitors Specific for LD2 or LD4 Motifs of Paxillin.
J.Mol.Biol., 427, 2015
|
|
8CT3
 
 | | Local refinement of band3-I transmembrane region from class 2 of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-13 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3M8E
 
 | | Protein structure of Type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Ni, L, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2BJQ
 
 | | Crystal structure of the nematode sperm cell motility protein MFP2 | | Descriptor: | MFP2A | | Authors: | Grant, R.P, Buttery, S.M, Ekman, G.C, Roberts, T.M, Stewart, M. | | Deposit date: | 2005-02-07 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Mfp2 and its Function in Enhancing Msp Polymerization in Ascaris Sperm Amoeboid Motility
J.Mol.Biol., 347, 2005
|
|
4XGZ
 
 | | Crystal structure of human paxillin LD2 motif in complex with Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Nocula-Lugowska, M, Lugowski, M, Salgia, R, Kossiakoff, A.A. | | Deposit date: | 2015-01-04 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering Synthetic Antibody Inhibitors Specific for LD2 or LD4 Motifs of Paxillin.
J.Mol.Biol., 427, 2015
|
|
8BT6
 
 | |
4XRE
 
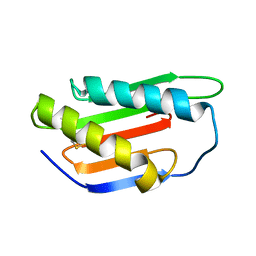 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
4Y92
 
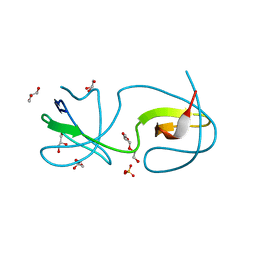 | |
2P27
 
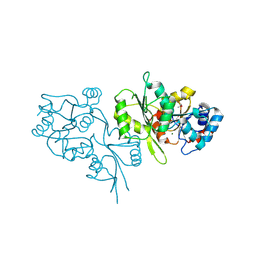 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
6T58
 
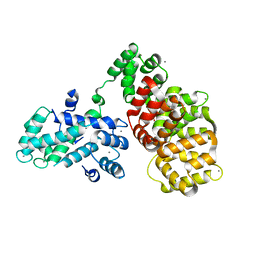 | | Structure determination of the transactivation domain of p53 in complex with S100A4 using annexin A2 as a crystallization chaperone | | Descriptor: | CALCIUM ION, Cellular tumor antigen p53,Protein S100-A4,Protein S100-A4,Annexin A2, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Nyitray, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure Determination of the Transactivation Domain of p53 in Complex with S100A4 Using Annexin A2 as a Crystallization Chaperone.
Structure, 28, 2020
|
|
2KR3
 
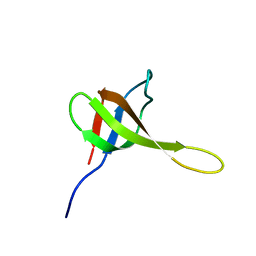 | | Solution structure of SHA-D | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Khristoforov, V.S, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, Y.A, Gushchina, L.V, Filimonov, V.V, Kutyshenko, V.P. | | Deposit date: | 2009-12-03 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Chimeric SHA-D domain "SH3-Bergerac": 3D structure and dynamics studies
Russ.J.Bioorganic Chem., 36, 2010
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
3M8K
 
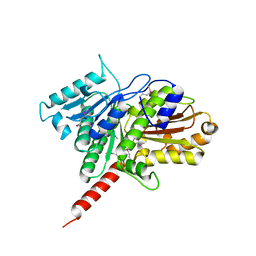 | | Protein structure of type III plasmid segregation TubZ | | Descriptor: | FtsZ/tubulin-related protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-18 | | Release date: | 2010-07-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M89
 
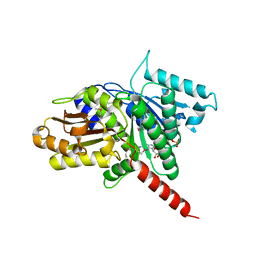 | | Structure of TubZ-GTP-g-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, FtsZ/tubulin-related protein | | Authors: | Ni, L, Xu, W, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M8F
 
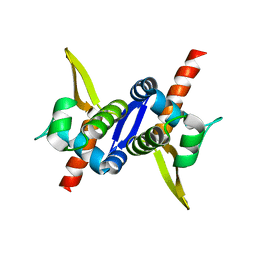 | | Protein structure of type III plasmid segregation TubR mutant | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2OYC
 
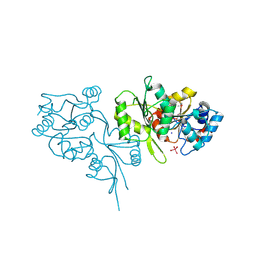 | | Crystal structure of human pyridoxal phosphate phosphatase | | Descriptor: | Pyridoxal phosphate phosphatase, SODIUM ION, TUNGSTATE(VI)ION | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
6X2V
 
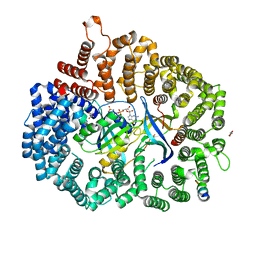 | | Crystal Structure of PKI(DE)NES peptide bound to CRM1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2O
 
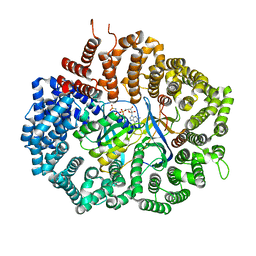 | | Crystal Structure of unliganded CRM1(E571K)-Ran-RanBP1 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2X
 
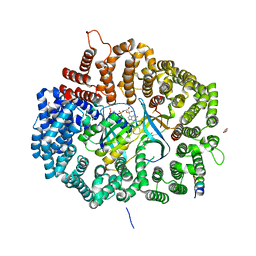 | | Crystal Structure of Mek1NES peptide bound to CRM1(E571K) | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2R
 
 | | Crystal Structure of the 4E-TNES peptide bound to CRM1 | | Descriptor: | Eukaryotic translation initiation factor 4E transporter, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2Y
 
 | |
2RD0
 
 | | Structure of a human p110alpha/p85alpha complex | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations.
Science, 318, 2007
|
|
