8AO3
 
 | | Specific covalent inhibitor of ERK2 | | Descriptor: | 2-chloranyl-~{N}-(1~{H}-indazol-5-ylmethyl)ethanamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO2
 
 | | Specific covalent inhibitor (3) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8ANS
 
 | | Crystal structure of D1228V c-MET bound by compound 1. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, GLYCEROL, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8ANF
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074359) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[3-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]propyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-05 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ANE
 
 | | Structure of the type I-G CRISPR effector | | Descriptor: | Cas7, RNA (66-MER) | | Authors: | Shangguan, Q, Graham, S, Sundaramoorthy, R, White, M.F. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the type I-G CRISPR effector.
Nucleic Acids Res., 50, 2022
|
|
8AN8
 
 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8AN6
 
 | |
8AN5
 
 | | MenAT1 toxin-antitoxin complex (rv0078a-rv0078b) from Mycobacterium tuberculosis H37Rv | | Descriptor: | Bacterial toxin, Conserved protein | | Authors: | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
8AMX
 
 | | AQP7 dimer of tetramers_D4 | | Descriptor: | Aquaporin-7 | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8AMW
 
 | | AQP7 dimer of tetramers_C1 | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8AMT
 
 | | OBD-RepB pMV158 domain | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Machon, C, Amodio, J, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8AMR
 
 | | Coiled-coil domain of human TRIM3 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Tripartite motif-containing protein 3 | | Authors: | Perez-Borrajero, C, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8AMO
 
 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMH
 
 | |
8AM8
 
 | |
8AM6
 
 | |
8AM3
 
 | |
8AM2
 
 | | Human butyrylcholinesterase in complex with 2,2'-(((1E,1'E)-(2-phenylpyrimidine-4,6-diyl)bis(methaneylylidene))bis(hydrazin-1-yl-2-ylidene))bis(N,N,N-trimethyl-2-oxoethan-1-aminium) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Dias, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Grid-Type Quaternary Metallosupramolecular Compounds Inhibit Human Cholinesterases through Dynamic Multivalent Interactions.
Chembiochem, 23, 2022
|
|
8AM1
 
 | | Human butyrylcholinesterase in complex with zinc and N,N,N-trimethyl-2-oxo-2-(2-(pyridin-2-ylmethylene)hydrazineyl)ethan-1-aminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Dias, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Grid-Type Quaternary Metallosupramolecular Compounds Inhibit Human Cholinesterases through Dynamic Multivalent Interactions.
Chembiochem, 23, 2022
|
|
8AM0
 
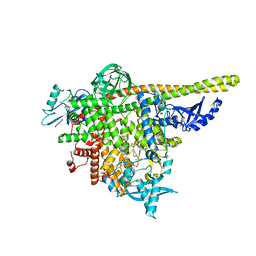 | | Crystal structure of human T1061E PI3Kalpha in complex with its regulatory subunit and the inhibitor GDC-0077 (Inavolisib) | | Descriptor: | (2R)-2-[[2-[(4S)-4-[bis(fluoranyl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl]amino]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Goncalves, M, Johnson, J.L, Roewer, K.M. | | Deposit date: | 2022-08-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Epinephrine inhibits PI3K alpha via the Hippo kinases.
Cell Rep, 42, 2023
|
|
8ALX
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, AMINOMETHYLAMIDE, ACETATE ION, ... | | Authors: | Rodriguez, I, Grudnik, P, Holak, T, Magiera-Mularz, K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biological characterization of pAC65, a macrocyclic peptide that blocks PD-L1 with equivalent potency to the FDA-approved antibodies.
Mol Cancer, 22, 2023
|
|
8ALS
 
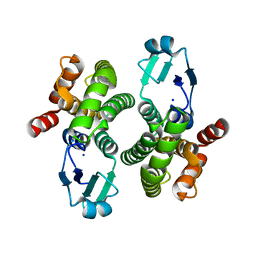 | |
8ALO
 
 | | Heterodimer formation of sensory domains of Vibrio cholerae regulators ToxR and ToxS | | Descriptor: | Cholera toxin transcriptional activator, Transmembrane regulatory protein ToxS | | Authors: | Gubensaek, N, Sagmeister, T, Pavkov-Keller, T, Zangger, K, Buhlheller, C, Wagner, G.E. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Vibrio cholerae's ToxRS bile sensing system.
Elife, 12, 2023
|
|
8ALN
 
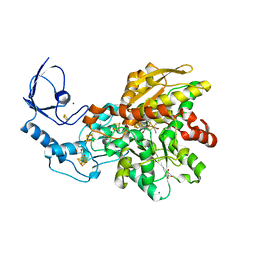 | | CO-bound [FeFe]-hydrogenase I from Clostridium pasteurianum (CpI) at 1.34 Angstrom | | Descriptor: | Binuclear [FeFe], di(thiomethyl)amine, carbon monoxide, ... | | Authors: | Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2022-08-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Cyanide Binding to [FeFe]-Hydrogenase Stabilizes the Alternative Configuration of the Proton Transfer Pathway.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
