424D
 
 | | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3') | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5CYY
 
 | |
3KC1
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 19a | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(7-carbamoyl-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4ELM
 
 | | Crystal structure of the mouse CD1d-lysosulfatide-Hy19.3 TCR complex | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl 3-O-sulfo-beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Maricic, I, Wang, J, Mac, T.T, Iyer, P, Kumar, V, Zajonc, D.M. | | Deposit date: | 2012-04-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens.
Nat.Immunol., 13, 2012
|
|
4J12
 
 | | monomeric Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, human Fc fragment | | Authors: | Ishino, T, Wang, M, Mosyak, L, Tam, A, Duan, W, Svenson, K, Joyce, A, O'Hara, D, Lin, L, Somers, W, Kriz, R. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Monomeric Fc Domain Modality by N-Glycosylation for the Half-life Extension of Biotherapeutics.
J.Biol.Chem., 288, 2013
|
|
1KJ2
 
 | | Murine Alloreactive ScFv TCR-Peptide-MHC Class I Molecule Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Allogeneic H-2Kb MHC Class I Molecule, Beta-2 microglobulin, ... | | Authors: | Reiser, J.-B, Gregoire, C, Darnault, C, Mosser, T, Guimezanes, A, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Mazza, G, Malissen, B, Housset, D. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex.
Immunity, 16, 2002
|
|
4JF7
 
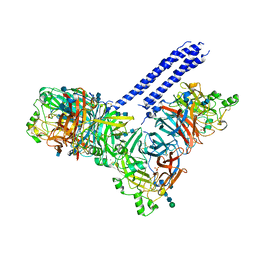 | | Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Welch, B.D, Yuan, P, Bose, S, Kors, C.A, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5018 Å) | | Cite: | Structure of the Parainfluenza Virus 5 (PIV5) Hemagglutinin-Neuraminidase (HN) Ectodomain.
Plos Pathog., 9, 2013
|
|
4RE6
 
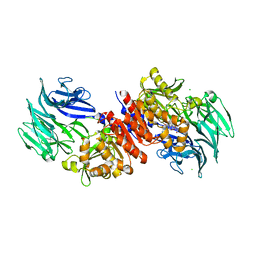 | | Acylaminoacyl peptidase complexed with a chloromethylketone inhibitor | | Descriptor: | Acylamino-acid-releasing enzyme, CHLORIDE ION, N-[(benzyloxy)carbonyl]glycyl-N-[(2S,3R)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]glycinamide | | Authors: | Menyhard, D.K, Orgovan, Z, Szeltner, Z, Szamosi, I, Harmat, V. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R1D
 
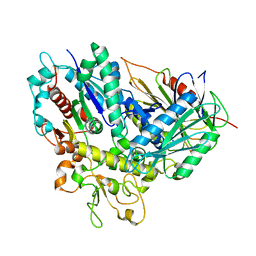 | | The crystal structure of Tle4-Tli4 complex | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Lu, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The structural basis of the Tle4-Tli4 complex reveals the self-protection mechanism of H2-T6SS in Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R36
 
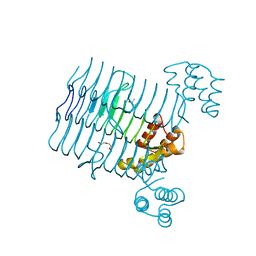 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1VSG
 
 | |
6KV5
 
 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
3NBE
 
 | | Clitocybe nebularis ricin B-like lectin (CNL) in complex with N,N'-diacetyllactosediamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-2-azidoethyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Ricin B-like lectin, SULFATE ION | | Authors: | Renko, M, Pohleven, J, Sabotic, J, Kos, J, Turk, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N'-diacetyllactosediamine (GalNAc beta 1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis
J.Biol.Chem., 287, 2012
|
|
4LKK
 
 | | The structure of hemagglutinin L226Q mutant (H3 numbering) from a avian-origin H7N9 influenza virus (A/Anhui/1/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, hemagglutinin | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-07-07 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses
Science, 342, 2013
|
|
4LAJ
 
 | | Crystal structure of HIV-1 YU2 envelope gp120 glycoprotein in complex with CD4-mimetic miniprotein, M48U1, and llama single-domain, broadly neutralizing, co-receptor binding site antibody, JM4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Heavy Chain-Only IgG2b Llama Antibody Effects Near-Pan HIV-1 Neutralization by Recognizing a CD4-Induced Epitope That Includes Elements of Coreceptor- and CD4-Binding Sites.
J.Virol., 87, 2013
|
|
2XJV
 
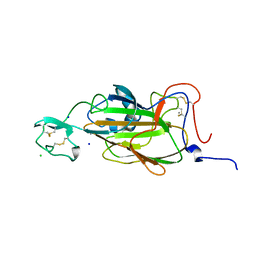 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae with mutation D201T in complex with calcium and glucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4KWU
 
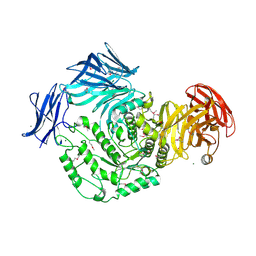 | | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e in complex with alpha-D-glucose, beta-D-glucose, magnesium and calcium | | Descriptor: | CALCIUM ION, Lmo2446 protein, MAGNESIUM ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e in complex with alpha-D-glucose, beta-D-glucose, magnesium and calcium
To be Published
|
|
1H1B
 
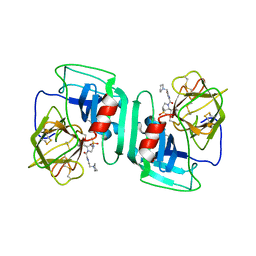 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
5AFB
 
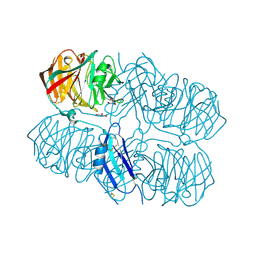 | | Crystal structure of the Latrophilin3 Lectin and Olfactomedin Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jackson, V.A, del Toro, D, Carrasquero, M, Roversi, P, Harlos, K, Klein, R, Seiradake, E. | | Deposit date: | 2015-01-21 | | Release date: | 2015-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis of Latrophilin-Flrt Interaction.
Structure, 23, 2015
|
|
7MM1
 
 | | PTP1B in complex with TCS401 by Native S-SAD at Room Temperature | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Dalton, K.M, Hekstra, D.R. | | Deposit date: | 2021-04-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Native SAD phasing at room temperature.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4AIC
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin, manganese and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose- 5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
4RE5
 
 | | Acylaminoacyl peptidase complexed with a chloromethylketone inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Acylamino-acid-releasing enzyme, ... | | Authors: | Menyhard, D.K, Orgovan, Z, Szeltner, Z, Szamosi, I, Harmat, V. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3KIF
 
 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib1-B7-18 and Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin, SULFATE ION | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
