3LSM
 
 | | Pyranose 2-oxidase H167A mutant with flavin N(5) sulfite adduct | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, DODECAETHYLENE GLYCOL, Pyranose 2-oxidase, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | H-bonding and positive charge at the N5/O4 locus are critical for covalent flavin attachment in trametes pyranose 2-oxidase.
J.Mol.Biol., 402, 2010
|
|
3LSN
 
 | | Crystal structure of putative geranyltranstransferase from PSEUDOMONAS fluorescens PF-5 complexed with magnesium | | Descriptor: | Geranyltranstransferase, MAGNESIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of putative geranyltranstransferase from Pseudomonas fluorescens PF-5 complexed with magnesium
To be Published
|
|
3LSO
 
 | |
3LSP
 
 | | Crystal Structure of DesT bound to desCB promoter and oleoyl-CoA | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*TP*CP*GP*AP*GP*TP*CP*AP*AP*CP*AP*AP*GP*CP*GP*TP*TP*CP*AP*CP*TP*GP*AP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*AP*TP*CP*AP*GP*TP*GP*AP*AP*CP*GP*CP*TP*TP*GP*TP*TP*GP*AP*CP*TP*CP*GP*AP*TP*TP*G)-3'), DesT, ... | | Authors: | Miller, D.J, White, S.W. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for the transcriptional regulation of membrane lipid homeostasis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LSQ
 
 | |
3LSR
 
 | |
3LSS
 
 | |
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | Descriptor: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3LSU
 
 | |
3LSV
 
 | |
3LSW
 
 | | Aniracetam bound to the ligand binding domain of GluA3 | | Descriptor: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, GLUTAMIC ACID, GluA2 S1S2 domain, ... | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-02-13 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
3LSX
 
 | | Piracetam bound to the ligand binding domain of GluA3 | | Descriptor: | 2-(2-oxopyrrolidin-1-yl)acetamide, GLUTAMIC ACID, GluA3 S1S2 domain, ... | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-02-13 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
3LSY
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T0 | | Descriptor: | 3-hydroxy-4-phenoxybenzaldehyde, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Bhargav, S.P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-02-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3LSZ
 
 | | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides
To be Published
|
|
3LT0
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T1 | | Descriptor: | 4-(2,4-dichlorophenoxy)-3-hydroxybenzaldehyde, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Bhargav, S.P, Sankaran, B, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-02-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3LT1
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T2 | | Descriptor: | 5-(chloromethyl)-2-(2,4-dichlorophenoxy)phenol, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Bhargav, S.P, Sankaran, B, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-02-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3LT2
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T3 | | Descriptor: | 2-(2,4-dichlorophenoxy)-5-(hydroxymethyl)phenol, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Bhargav, S.P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-02-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3LT3
 
 | | Crystal structure of Rv3671c from M. tuberculosis H37Rv, Ser343Ala mutant, inactive form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2010-02-14 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
3LT4
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant PB4 | | Descriptor: | 2-(2-amino-4-chlorophenoxy)-5-chlorophenol, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Bhargav, S.P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3LT5
 
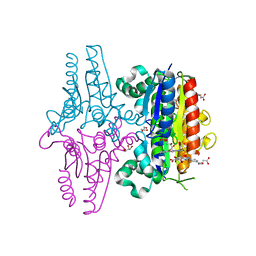 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | Descriptor: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
3LT6
 
 | |
3LT7
 
 | |
3LT8
 
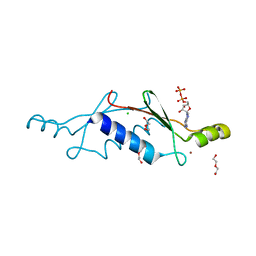 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding, crystallized in the presence of 100 mM ATP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LT9
 
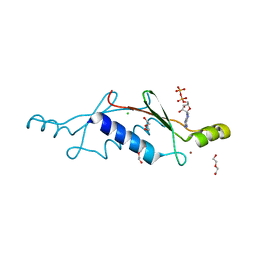 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LTA
 
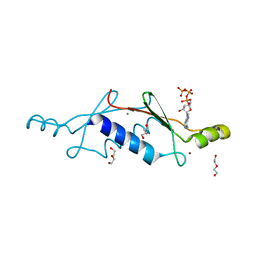 | | Crystal structure of a non-biological ATP binding protein with a TYR-PHE mutation within the ligand binding domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
