4UGP
 
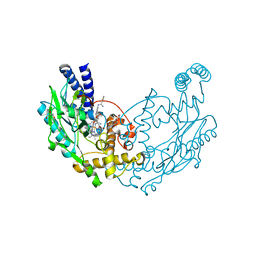 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N',N'-(((2R)-3-aminopropane-1,2-diyl)bis(oxymethanediylbenzene-3,1- diyl))dithiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4FW3
 
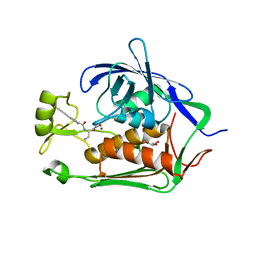 | |
4H2V
 
 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with glycylated carrier protein | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
7L7L
 
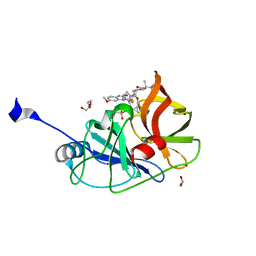 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-129 | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
6NQY
 
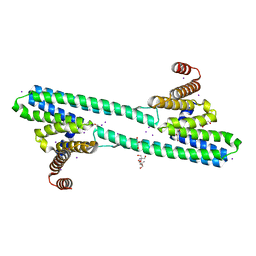 | | Flagellar protein FcpA from Leptospira biflexa / ab-centered monoclinic form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Flagellar coiling protein A, ... | | Authors: | Mechaly, A, Larrieux, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete.
Elife, 9, 2020
|
|
8HHP
 
 | |
1SUE
 
 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
6L30
 
 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2 | | Authors: | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | Deposit date: | 2019-10-07 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6J56
 
 | | Crystal structure of Myosin VI CBD in complex with Tom1 MBM | | Descriptor: | Peptide from Target of Myb protein 1, Unconventional myosin-VI | | Authors: | Hu, S, Pan, L. | | Deposit date: | 2019-01-10 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering.
Nat Commun, 10, 2019
|
|
8I2N
 
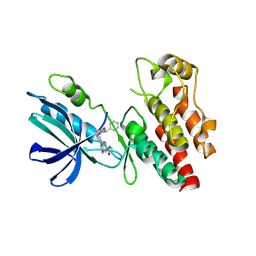 | | The RIPK1 kinase domain in complex with QY7-2B compound | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-methyl-1-[4-[[[1-methyl-5-(phenylmethyl)pyrazol-3-yl]carbonylamino]methyl]phenyl]benzimidazole-5-carboxamide | | Authors: | Gong, X.Y, Li, Y, Meng, H.Y, Pan, L.F. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The RIPK1 kinase domain in complex with QY7-2B compound
To Be Published
|
|
8PZK
 
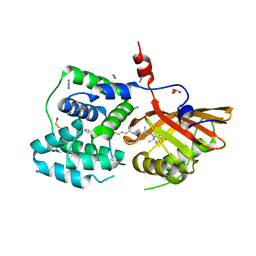 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8QIJ
 
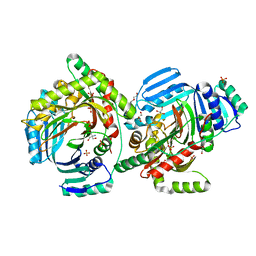 | | Crystallographic Structure of a Salicylate Synthase from M. abscessus (Mab-SaS) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cassetta, A, Covaceuszach, S, Tomaiuolo, M, Meneghetti, F, Villa, S, Mori, M, Chiarelli, L.R, Mangiatordi, G.F. | | Deposit date: | 2023-09-12 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural basis for specific inhibition of salicylate synthase from Mycobacterium abscessus.
Eur.J.Med.Chem., 265, 2024
|
|
8QAX
 
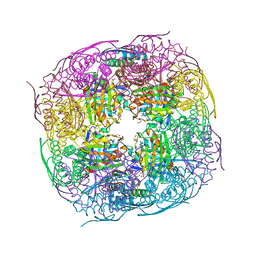 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, GOL and TRS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
7NPA
 
 | |
4PXS
 
 | |
8QZF
 
 | | Heme-domain BM3 mutant T268E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
5NCW
 
 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
8BJH
 
 | | chimera of the inactive ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, with the double mutation K3528M and K3535I, fused to a proline-Rich-Domain (PRD) and profilin, bound to Latrunculin B-ADP-Mg-actin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BJI
 
 | | chimera of ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo fused to a proline-Rich-Domain (PRD) and profilin, bound to ADP-Mg-actin and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BJJ
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to ATP-Mg-actin, human profilin 1 and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
1T79
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxLW motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxLW motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-08 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
7M00
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 13c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
1T74
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a WxxLF motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T7F
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a LxxLL motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, LxxLL motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T7M
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxYF motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
