5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
5DQF
 
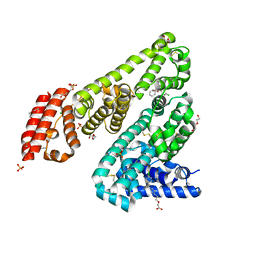 | | Horse Serum Albumin (ESA) in complex with Cetirizine | | Descriptor: | (2-{4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, (2-{4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, CHLORIDE ION, ... | | Authors: | Handing, K.B, Shabalin, I.G, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site.
Mol.Immunol., 71, 2016
|
|
2YI1
 
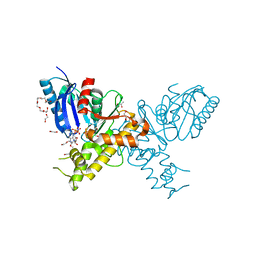 | | Crystal structure of N-Acetylmannosamine kinase in complex with N- acetyl mannosamine 6-phosphate and ADP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
8DSV
 
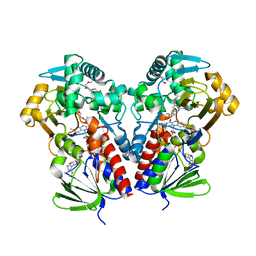 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
3RHJ
 
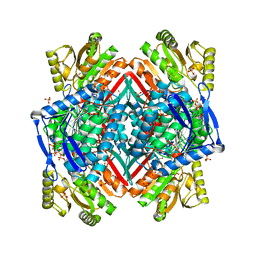 | |
3RHL
 
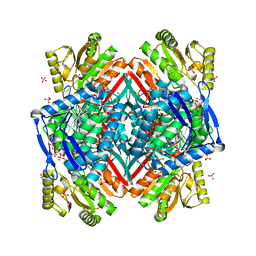 | |
3RHR
 
 | |
8DSW
 
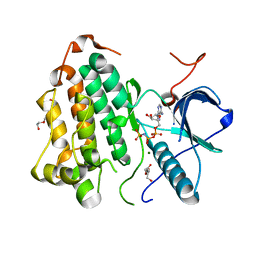 | |
8E0G
 
 | | Re-refined model of active mu-opioid receptor (PDB 5c1m) as an adduct with BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Munro, T.A. | | Deposit date: | 2022-08-09 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reanalysis of a mu opioid receptor crystal structure reveals a covalent adduct with BU72.
Bmc Biol., 21, 2023
|
|
8DYD
 
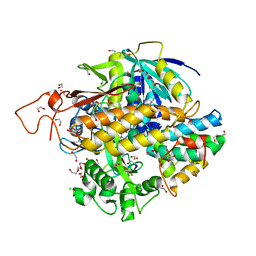 | | Crystal structure of human SDHA-SDHAF2-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
8F46
 
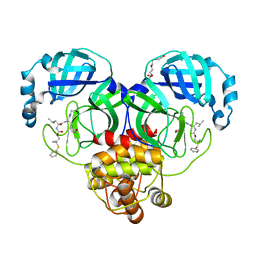 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor (cyano warhead) | | Descriptor: | 3C-like proteinase, N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide, TETRAETHYLENE GLYCOL | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8F44
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8F8O
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids
To Be Published
|
|
3OAB
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
2V3V
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-22 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
6ARR
 
 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with cesium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CESIUM ION, CHLORIDE ION, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
3RHO
 
 | |
6ART
 
 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with cesium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CESIUM ION, CHLORIDE ION, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
2VPZ
 
 | | Polysulfide reductase native structure | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration
Nat.Struct.Mol.Biol., 15, 2008
|
|
3SDA
 
 | |
7CJS
 
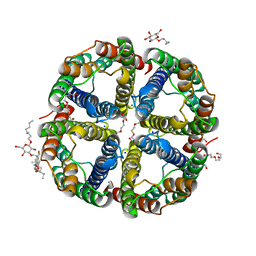 | | structure of aquaporin | | Descriptor: | Aquaporin NIP2-1, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Saitoh, Y, Ma, J.F, Suga, M. | | Deposit date: | 2020-07-13 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for high selectivity of a rice silicon channel Lsi1.
Nat Commun, 12, 2021
|
|
6ARL
 
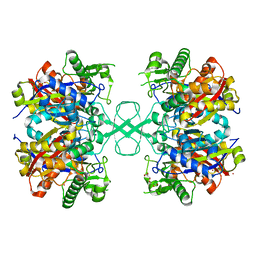 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with rubidium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
7BJL
 
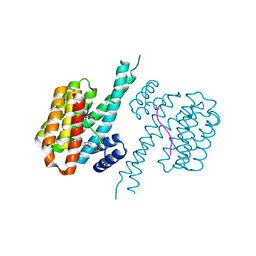 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-142 | | Descriptor: | 14-3-3 protein sigma, 3-[(3~{S})-3-methoxypiperidin-1-yl]-4-nitro-benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJW
 
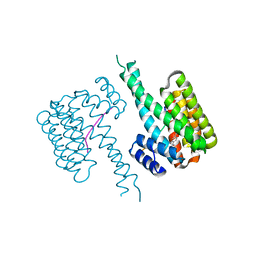 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-154 | | Descriptor: | 14-3-3 protein sigma, 4-nitro-3-(4-oxidanylpiperidin-1-yl)benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIQ
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-180 | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{R})-2-methyl-3,4-dihydro-2~{H}-quinolin-1-yl]sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
