3CVP
 
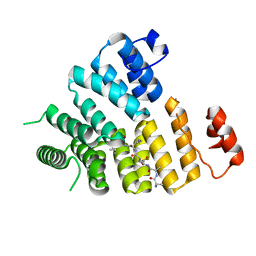 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (10-SKL) | | Descriptor: | 10-SKL PTS1 peptide Ac-GTLSNRASKL, Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVN
 
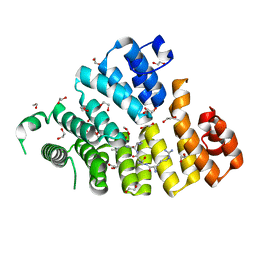 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor, T. brucei GAPDH PTS1 peptide Ac-DRDAAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3RU6
 
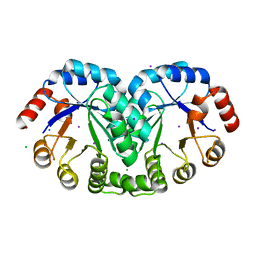 | | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, IODIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3OTR
 
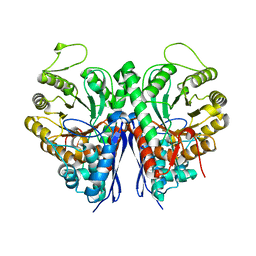 | | 2.75 Angstrom Crystal Structure of Enolase 1 from Toxoplasma gondii | | Descriptor: | CHLORIDE ION, Enolase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Shuvalova, L, Halavaty, A, Ngo, H, Tomavo, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of bradyzoite-specific enolase from Toxoplasma gondii reveals insights into its dual cytoplasmic and nuclear functions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3OPZ
 
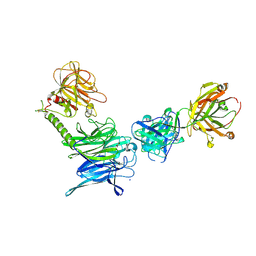 | | Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, Trans-sialidase, ... | | Authors: | Larrieux, N, Muia, R, Campetella, O, Buschiazzo, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Plos Pathog., 8, 2012
|
|
3OO2
 
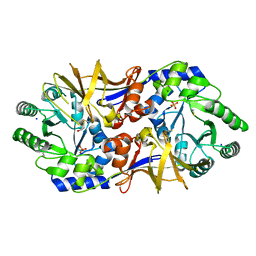 | | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Alanine racemase 1, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3SG1
 
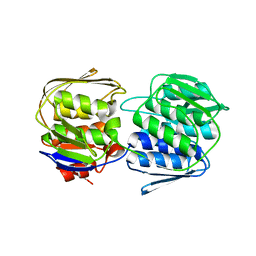 | | 2.6 Angstrom Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurA1) from Bacillus anthracis | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | | Authors: | Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurA1) from Bacillus anthracis.
TO BE PUBLISHED
|
|
6X5Q
 
 | |
6X5G
 
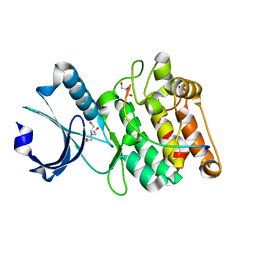 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and LRRC7 inhibitory domain | | Descriptor: | BICINE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
5E5E
 
 | |
5E63
 
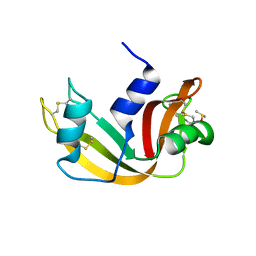
 | | K262A mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5DP9
 
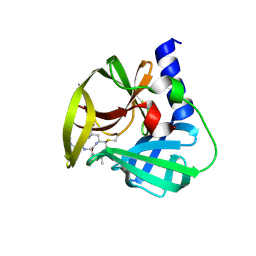 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
4W1V
 
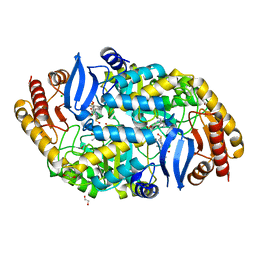 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a thiazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
6T37
 
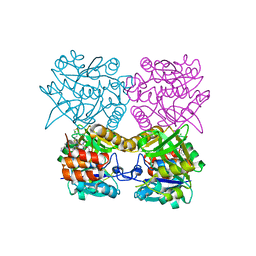 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
4WD2
 
 | |
5DY5
 
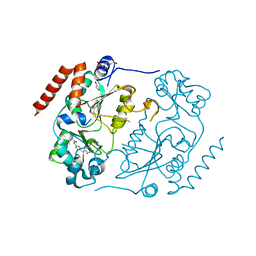 | | Crystal structure of human Sirt2 in complex with a SirReal probe fragment | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Development of an Affinity Probe for Sirtuin 2.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E9R
 
 | |
5E67
 
 | | K103A/K262A double mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA bottom strand, ... | | Authors: | Shen, B.W, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5EDK
 
 | | Crystal structure of prothrombin deletion mutant residues 146-167 ( Form II ). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
6TQG
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
5DW8
 
 | |
6G5U
 
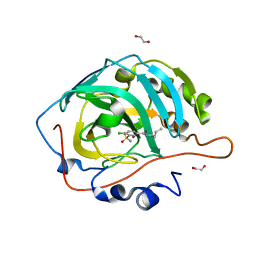 | | Crystal structure of human carbonic anhydrase isozyme XIII with N-butyl-2,4-dichloro-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Carbonic anhydrase 13, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2018-03-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of two-tail compounds with rotationally fixed benzenesulfonamide ring as inhibitors of carbonic anhydrases.
Eur J Med Chem, 156, 2018
|
|
6TRJ
 
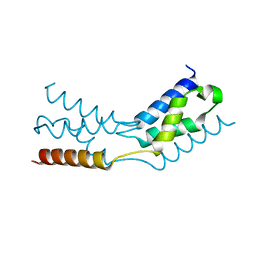 | | LEDGF/p75 IBD dimer | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Kugler, M, Brynda, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fine-tuning of the LEDGF/p75 interaction network by dimerization
Structure
|
|
6XE3
 
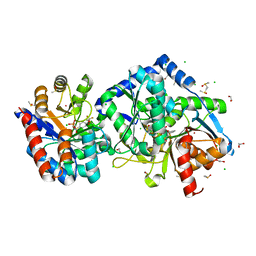 | | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site. | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-06-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site.
To be Published
|
|
6XQK
 
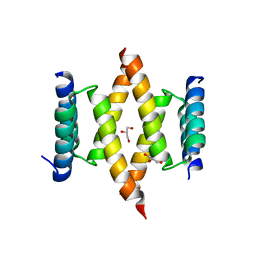 | | Crystal structure of the D/D domain of PKA from S. cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, cAMP-dependent protein kinase regulatory subunit | | Authors: | Larrieux, N, Gonzalez Bardeci, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of yeast regulatory subunit reveals key evolutionary insights into Protein Kinase A oligomerization.
J.Struct.Biol., 213, 2021
|
|
