4OS2
 
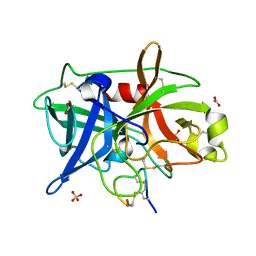 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK602 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4INB
 
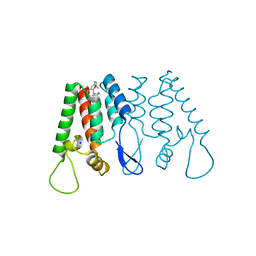 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With benzodiazepine Inhibitor | | Descriptor: | (3Z)-3-{[(2-methoxyethyl)amino]methylidene}-1-methyl-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, Gag protein, SODIUM ION | | Authors: | Coulombe, R. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monitoring Binding of HIV-1 Capsid Assembly Inhibitors Using (19) F Ligand-and (15) N Protein-Based NMR and X-ray Crystallography: Early Hit Validation of a Benzodiazepine Series.
Chemmedchem, 8, 2013
|
|
3QRN
 
 | |
3QYU
 
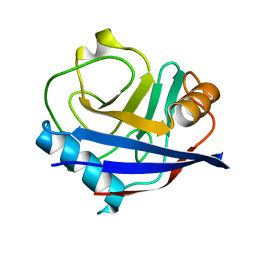 | | Crystal structure of human cyclophilin D at 1.54 A resolution at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F | | Authors: | Colliandre, L, Gelin, M, Labesse, G, Guichou, J.-F. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3QYZ
 
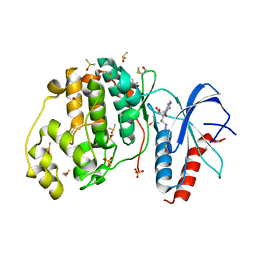 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2HG1
 
 | |
1WOB
 
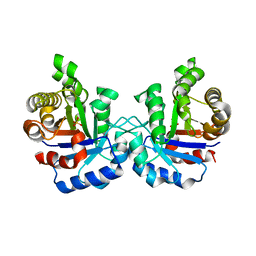 | | Structure of a loop6 hinge mutant of Plasmodium falciparum Triosephosphate Isomerase, W168F, complexed to sulfate | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Eaazhisai, K, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2004-08-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Unliganded and Inhibitor Complexes of W168F, a Loop6 Hinge Mutant of Plasmodium falciparum Triosephosphate Isomerase: Observation of an Intermediate Position of Loop6
J.Mol.Biol., 343, 2004
|
|
9EN6
 
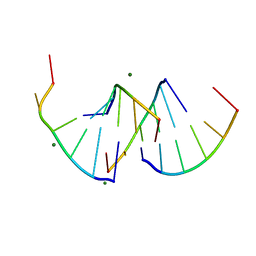 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Mateja-Pluta, M, Kiliszek, A. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.918 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
2Y4O
 
 | | Crystal Structure of PaaK2 in complex with phenylacetyl adenylate | | Descriptor: | 5'-O-[HYDROXY(PHENYLACETYL)PHOSPHORYL]ADENOSINE, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Law, A, Boulanger, M.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Defining a Structural and Kinetic Rationale for Paralogous Copies of Phenylacetate-Coa Ligases from the Cystic Fibrosis Pathogen Burkholderia Cenocepacia J2315.
J.Biol.Chem., 286, 2011
|
|
2Y43
 
 | |
9EO0
 
 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
8YBB
 
 | |
8EX1
 
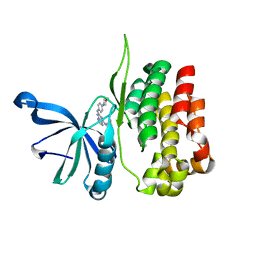 | |
8WWB
 
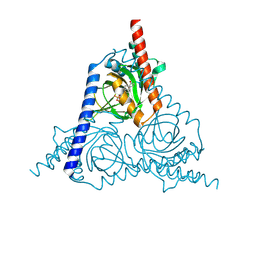 | |
8EX0
 
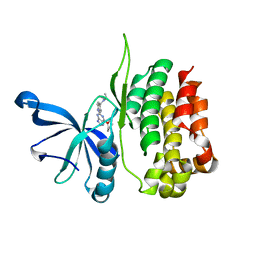 | |
8EX2
 
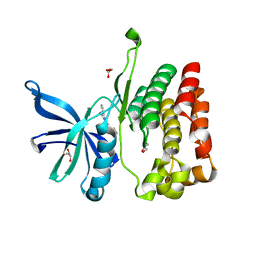 | |
8BIT
 
 | | Crystal structure of acyl-CoA synthetase from Metallosphaera sedula in complex with Coenzyme A and acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, COENZYME A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
5E7Q
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis | | Descriptor: | GLYCEROL, SULFATE ION, acyl-CoA synthetase | | Authors: | Osipiuk, J, Cuff, M.E, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-12 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
2JDX
 
 | |
3QCC
 
 | |
6ZW0
 
 | |
6ZVZ
 
 | |
7NA0
 
 | |
3QCE
 
 | |
1WOA
 
 | | Structure of the loop6 hinge mutant of Plasmodium falciparum Triosephosphate Isomerase, W168F, complexed with Glycerol-2-phosphate | | Descriptor: | 2-HYDROXY-1-(HYDROXYMETHYL)ETHYL DIHYDROGEN PHOSPHATE, Triosephosphate isomerase | | Authors: | Eaazhisai, K, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2004-08-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Unliganded and Inhibitor Complexes of W168F, a Loop6 Hinge Mutant of Plasmodium falciparum Triosephosphate Isomerase: Observation of an Intermediate Position of Loop6
J.Mol.Biol., 343, 2004
|
|
