5RD6
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 28 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDN
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 47 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4B2H
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | 3-[7,8-dimethyl-2,4-bis(oxidanylidene)benzo[g]pteridin-10-yl]propylcarbamic acid, CHLORIDE ION, DODECIN, ... | | Authors: | Yu, Y, Heidel, B, Parapugna, T.L, Wenderhold-Reeb, S, Song, B, Schoenherr, H, Grininger, M, Noell, G. | | Deposit date: | 2012-07-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Flavoprotein Dodecin as a Redox Probe for Electron Transfer Through DNA.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4B30
 
 | |
5UXY
 
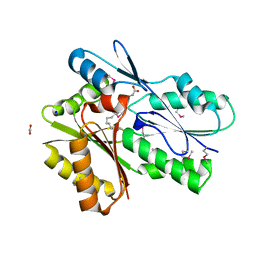 | | The crystal structure of a DegV family protein from Eubacterium eligens loaded with heptadecanoic acid to 1.80 Angstrom resolution (ALTERNATIVE REFINEMENT OF PDB 3FDJ with HEPTADECANOIC acid) | | Descriptor: | ACETIC ACID, DegV family protein, SODIUM ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-23 | | Release date: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
5VB5
 
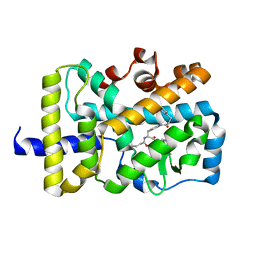 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an inverse agonist and SRC2 peptide | | Descriptor: | N-[(2R)-3-(4-{[3-(4-chlorophenyl)propanoyl]amino}phenyl)-1-(4-methylpiperidin-1-yl)-1-oxopropan-2-yl]-4-methylpentanamide, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5V9F
 
 | |
5V8E
 
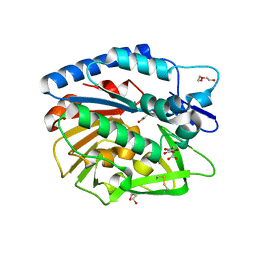 | | Structure of Bacillus cereus PatB1 | | Descriptor: | Bacillus cereus PatB1, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
5V97
 
 | |
5VFZ
 
 | |
1H18
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate | | Descriptor: | FORMATE ACETYLTRANSFERASE 1, L-TREITOL, PYRUVIC ACID, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1H16
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate and CoA | | Descriptor: | COENZYME A, FORMATE ACETYLTRANSFERASE 1, L-TREITOL, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
5VJO
 
 | |
1H17
 
 | |
5VI0
 
 | |
6QWU
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 5.5 with Mn2+ and CoA. | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
4TVT
 
 | | New ligand for thaumatin discovered using acoustic high throughput screening | | Descriptor: | ASCORBIC ACID, L(+)-TARTARIC ACID, SODIUM ION, ... | | Authors: | Teplitsky, E, Joshi, K, Ericson, D.L, Scalia, A, Mullen, J.D, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
6QLL
 
 | | Crystal structure of F181H UbiX in complex with FMN and dimethylallyl monophosphate | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dimethylallyl monophosphate, Flavin prenyltransferase UbiX, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry.
Nat Commun, 10, 2019
|
|
5ZOU
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH6 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5E1I
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
6QN4
 
 | |
5DMU
 
 | | Structure of the NHEJ polymerase from Methanocella paludicola | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NHEJ Polymerase, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
5ZMW
 
 | | Crystal structure of the E309Q mutant of SR Ca2+-ATPase in E2(TG) | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Ogawa, H, Hirata, A, Tsueda, J, Toyoshima, C. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of the E2 to E1 transition in Ca2+pump revealed by crystal structures of gating residue mutants.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5DRG
 
 | | Green/cyan WasCFP at pH 10.0 | | Descriptor: | GLYCEROL, Green/cyan WasCFP_pH10 at pH 10.0, SODIUM ION | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
4TW9
 
 | | Difluoro-dioxolo-benzoimidazol-benzamides as potent inhibitors of CK1delta and epsilon with nanomolar inhibitory activity on cancer cell proliferation | | Descriptor: | CHLORIDE ION, Casein kinase I isoform delta, N-(2,2-difluoro-5H-[1,3]dioxolo[4,5-f]benzimidazol-6-yl)-2-{[2-(trifluoromethoxy)benzoyl]amino}-1,3-thiazole-4-carboxamide, ... | | Authors: | Richter, J, Bischof, J, Zaja, M, Kohlhof, H, Othersen, O, Vitt, D, Alscher, V, Pospiech, I, Garcia-Reyes, B, Berg, S, Leban, J, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Difluoro-dioxolo-benzoimidazol-benzamides As Potent Inhibitors of CK1 delta and epsilon with Nanomolar Inhibitory Activity on Cancer Cell Proliferation.
J.Med.Chem., 57, 2014
|
|
