9J19
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 3C-like proteinase nsp5 | | Authors: | Singh, A, Jangid, K, Dhaka, P, Tomar, S, Kumar, P. | | Deposit date: | 2024-08-04 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into the Main Protease (Mpro) Dimer Interface Destabilization Inhibitor: Unveiling New Therapeutic Avenues against SARS-CoV-2.
Biochemistry, 64, 2025
|
|
7VQM
 
 | | GH2 beta-galacturonate AqGalA in complex with galacturonide | | Descriptor: | CHLORIDE ION, GH2 beta-galacturonate AqGalA, beta-D-galactopyranuronic acid | | Authors: | Yang, J. | | Deposit date: | 2021-10-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Basis of a Marine Bacterial Glycoside Hydrolase Family 2 beta-Glycosidase with Broad Substrate Specificity
Appl.Environ.Microbiol., 88, 2022
|
|
8K5W
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
9J4Z
 
 | |
9J50
 
 | |
7WPK
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with L-Met | | Descriptor: | METHIONINE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPL
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPN
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPJ
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPM
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
8OKP
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_33 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},15~{R})-10,13,15-trimethyl-17-oxidanylidene-1,2,3,4,5,6,7,8,9,11,12,14,15,16-tetradecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OKT
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OMN
 
 | | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl VK4 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OKO
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_2 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},8~{S},9~{S},10~{R},13~{S},14~{S})-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OLA
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor VK4 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OMH
 
 | | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl (BS1982) | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{S},9~{S},13~{S},14~{S})-13-methyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OKJ
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_12 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{S},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OLK
 
 | |
8OMP
 
 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl AKI33 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},15~{R})-10,13,15-trimethyl-17-oxidanylidene-1,2,3,4,5,6,7,8,9,11,12,14,15,16-tetradecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OLM
 
 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl AKI1 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},5~{S},8~{S},13~{R},14~{R})-5,14-dimethyl-16-oxidanylidene-1,2,3,4,6,7,8,11,12,13,15,17-dodecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OMB
 
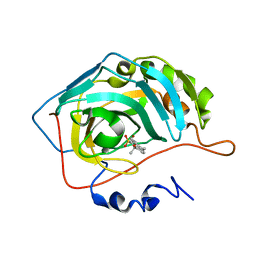 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OLI
 
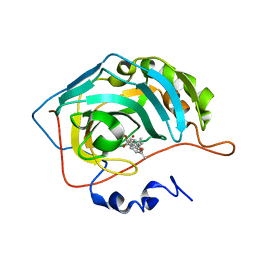 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl AKI_12 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{S},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OLF
 
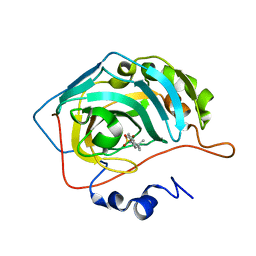 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor BS1982 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{S},9~{S},13~{S},14~{S})-13-methyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
7VE2
 
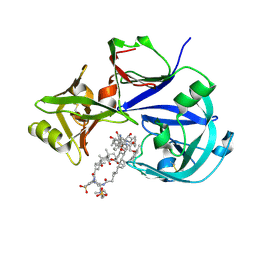 | | Crystal Structure of Lopinavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Plasmepsin II | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
