5CDR
 
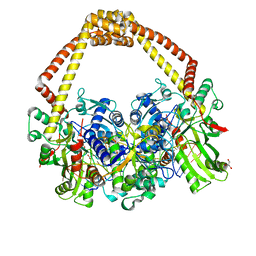 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDP
 
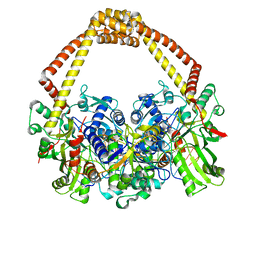 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
4P58
 
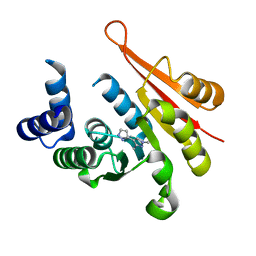 | | Crystal structure of mouse comt bound to an inhibitor | | Descriptor: | 1',3'-dimethyl-1H,1'H-3,4'-bipyrazole, Catechol O-methyltransferase | | Authors: | Lanier, M. | | Deposit date: | 2014-03-15 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A fragment-based approach to identifying S-adenosyl-l-methionine -competitive inhibitors of catechol O-methyl transferase (COMT).
J.Med.Chem., 57, 2014
|
|
3HIE
 
 | |
3GBS
 
 | | Crystal structure of Aspergillus oryzae cutinase | | Descriptor: | Cutinase 1 | | Authors: | Gosser, Y, Lu, Z, Alemu, G, Li, H, Kong, X, Liu, Z, Montclare, J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation.
J.Am.Chem.Soc., 131, 2009
|
|
5KX8
 
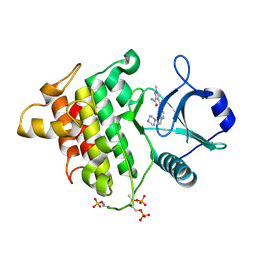 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(3-aminocarbonyl-1-methyl-pyrazol-4-yl)-5-piperazin-1-yl-pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1WCT
 
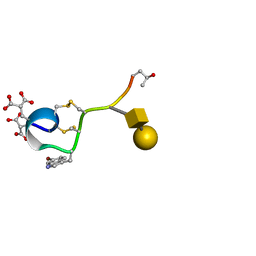 | | A NOVEL CONOTOXIN FROM CONUS TEXTILE WITH UNUSUAL POST-TRANSLATIONAL MODIFICATIONS REDUCES PRESYNAPTIC CALCIUM INFLUX, NMR, 1 STRUCTURE, GLYCOSYLATED PROTEIN | | Descriptor: | OMEGAC-TXIX, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Rigby, A.C, Hambe, B, Czerwiec, E, Baleja, J.D, Furie, B.C, Furie, B, Stenflo, J. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-08 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | A conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5KX7
 
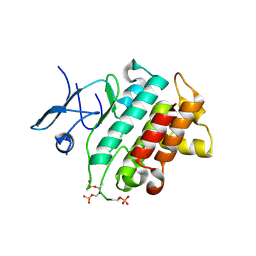 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(3-aminocarbonyl-1-methyl-pyrazol-4-yl)-6-(1-methylpyrazol-4-yl)pyridine-2-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8OLU
 
 | |
5M2N
 
 | |
5LDN
 
 | | A viral capsid:antibody complex | | Descriptor: | Hexon protein,hexon capsid, antibody | | Authors: | James, L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-antigen kinetics constrain intracellular humoral immunity.
Sci Rep, 6, 2016
|
|
5MN2
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2H9W
 
 | |
3NOS
 
 | | HUMAN ENDOTHELIAL NITRIC OXIDE SYNTHASE WITH ARGININE SUBSTRATE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ENDOTHELIAL NITRIC-OXIDE SYNTHASE, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Fischmann, T.O, Weber, P.C. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation.
Nat.Struct.Biol., 6, 1999
|
|
9CXR
 
 | | X-ray Crystallographic Structure of the Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 at Room Temperature | | Descriptor: | 1,2-ETHANEDIOL, Poly(hexamethylene adipamide) hydrolase, SODIUM ION, ... | | Authors: | Meilleur, F, Williams, A.N, Drufva, E.E, Parks, J.M, Chen, S.H, Michener, J.K. | | Deposit date: | 2024-07-31 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and characterization of substrate- and product-selective nylon hydrolases.
Biorxiv, 2024
|
|
6GFJ
 
 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
1DXS
 
 | |
3RDC
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combining 'dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9DYS
 
 | |
6M7W
 
 | |
8BRK
 
 | |
8BRL
 
 | |
3BJI
 
 | | Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 | | Descriptor: | Proto-oncogene vav, Ras-related C3 botulinum toxin substrate 1 precursor, ZINC ION | | Authors: | Chrencik, J.E, Brooun, A, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1.
J.Mol.Biol., 380, 2008
|
|
3ASW
 
 | |
8BP0
 
 | |
