4UTX
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3-nitro- propionylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 3-NITROPROPANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UUA
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3S-Z-amino- succinylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBAMOYLPHOSPHATE SYNTHETASE I, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4V0J
 
 | | The channel-block Ser202Glu, Thr104Lys double mutant of Stearoyl-ACP- Desaturase from Castor bean (Ricinus communis) | | Descriptor: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, CHLOROPLASTIC, FE (II) ION, ... | | Authors: | Moche, M, Guy, J, Whittle, E, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2014-09-17 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Half-of-the-Sites Reactivity of the Castor Delta9-18:0-Acp Desaturase.
Plant Physiol., 169, 2015
|
|
4WG0
 
 | |
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
7KTR
 
 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4QEX
 
 | | Crystal structure of PfEBA-175 RII in complex with a Fab fragment from inhibitory antibody R217 | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, Erythrocyte-binding antigen-175 | | Authors: | Chen, E, Paing, M.M, Salinas, N, Sim, B.K, Tolia, N.H. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-04 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Functional Basis for Inhibition of Erythrocyte Invasion by Antibodies that Target Plasmodium falciparum EBA-175.
Plos Pathog., 9, 2013
|
|
2RKX
 
 | | The 3D structure of chain D, cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Descriptor: | Cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Authors: | Tawfik, D, Khersonsky, O, Albeck, S, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-10-18 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kemp elimination catalysts by computational enzyme design.
Nature, 453, 2008
|
|
2RVD
 
 | |
6Y3M
 
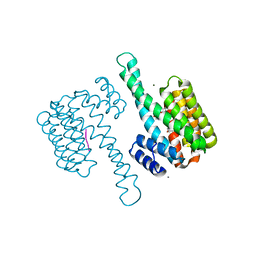 | | 14-3-3 Sigma in complex with phosphorylated ATPase peptide | | Descriptor: | 14-3-3 protein sigma, ATPase peptide, CALCIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3UKF
 
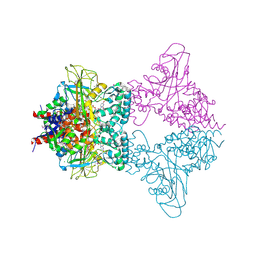 | |
3UKP
 
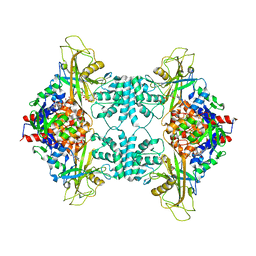 | |
3UKH
 
 | |
3UKL
 
 | |
5YBF
 
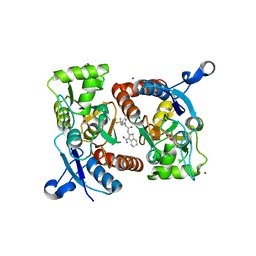 | | Crystal structure of the GluA2o LBD in complex with glutamate and HBT1 | | Descriptor: | 2-[2-[5-methyl-3-(trifluoromethyl)pyrazol-1-yl]ethanoylamino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
5YBG
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and LY451395 | | Descriptor: | ACETATE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
9B27
 
 | | Dia1 at the Barbed End of F-Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
8A2L
 
 | |
8A5N
 
 | |
8A2M
 
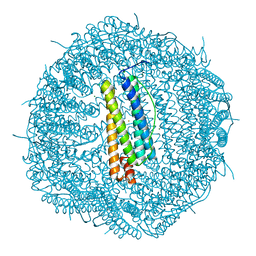 | |
5AEJ
 
 | | Crystal structure of human Gremlin-1 | | Descriptor: | GREMLIN-1, SULFATE ION | | Authors: | Kisonaite, M, Hyvonen, M. | | Deposit date: | 2015-08-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structure of Gremlin-1 and Analysis of its Interaction with Bmp-2.
Biochem.J., 473, 2016
|
|
1O07
 
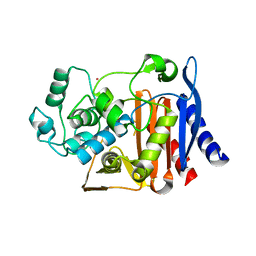 | | Crystal Structure of the complex between Q120L/Y150E mutant of AmpC and a beta-lactam inhibitor (MXG) | | Descriptor: | 2-(1-{2-[4-(2-ACETYLAMINO-PROPIONYLAMINO)-4-CARBOXY-BUTYRYLAMINO]-6-AMINO-HEXANOYLAMINO}-2-OXO-ETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, POTASSIUM ION | | Authors: | Meroueh, S.O, Minasov, G, Lee, W, Shoichet, B.K, Mobashery, S. | | Deposit date: | 2003-02-20 | | Release date: | 2003-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Aspects for Evolution of beta-Lactamases from Penicillin-Binding Proteins
J.Am.Chem.Soc., 125, 2003
|
|
7NWI
 
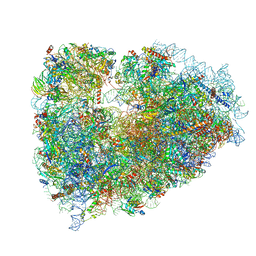 | | Mammalian pre-termination 80S ribosome with Empty-A site bound by Blasticidin S | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA+BlaS, 40S ribosomal protein S11, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7NWG
 
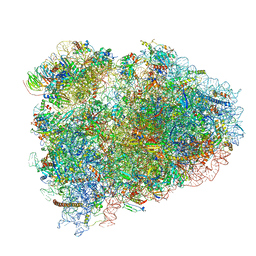 | | Mammalian pre-termination 80S ribosome with Hybrid P/E- and A/P-site tRNA's bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
6X18
 
 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
