5M2N
 
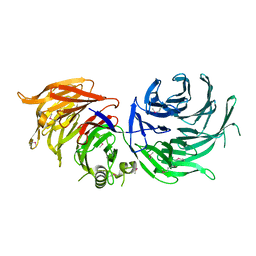 | |
1EO1
 
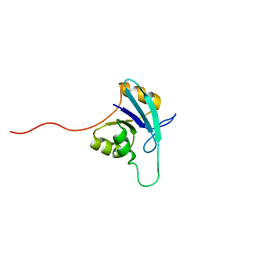 | |
5M34
 
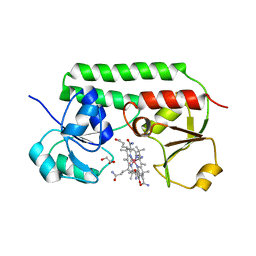 | | Structure of cobinamide-bound BtuF mutant W66Y, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
1EFC
 
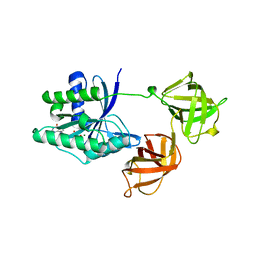 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
5M3E
 
 | | Macrodomain of Thermus aquaticus DarG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Appr-1-p processing domain protein, CHLORIDE ION | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
1EFV
 
 | |
5M3J
 
 | | Influenza B polymerase bound to four heptad repeats of serine 5 phosphorylated Pol II CTD | | Descriptor: | DNA-directed RNA polymerase subunit, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Lukarska, M, Pflug, A, Cusack, S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of an essential interaction between influenza polymerase and Pol II CTD.
Nature, 541, 2017
|
|
1EPJ
 
 | |
5LXX
 
 | | High-resolution structure of human collapsin response mediator protein 2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Myllykoski, M, Hensley, K, Kursula, P. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Collapsin response mediator protein 2: high-resolution crystal structure sheds light on small-molecule binding, post-translational modifications, and conformational flexibility.
Amino Acids, 49, 2017
|
|
1EPX
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ALDOLASE FROM L. MEXICANA | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Chudzik, D.M, Michels, P.A, de Walque, S, Hol, W.G.J. | | Deposit date: | 2000-03-29 | | Release date: | 2000-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of type 2 peroxisomal targeting signals in two trypanosomatid aldolases.
J.Mol.Biol., 300, 2000
|
|
5LZ4
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 5-[2-(4,4-dimethyl-2-oxidanylidene-pyrrolidin-1-yl)ethoxy]-2-fluoranyl-benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
1EQV
 
 | |
5LZJ
 
 | | Cholera toxin El Tor B-pentamer in complex with inhibitor Laura237 | | Descriptor: | (~{Z})-~{N}-[2-[(2~{R},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethyl]-3-(3,4,5-trimethoxyphenyl)prop-2-enamide, Cholera enterotoxin subunit B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Heggelund, J.E, Krengel, U. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Towards new cholera prophylactics and treatment: Crystal structures of bacterial enterotoxins in complex with GM1 mimics.
Sci Rep, 7, 2017
|
|
9GEJ
 
 | | Crystal structure of CREBBP bromodomain in complex with (2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione | | Descriptor: | 2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione, CREBBP, SULFATE ION | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione
To Be Published
|
|
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5M48
 
 | |
5M4D
 
 | | Alpha-amino epsilon-caprolactam racemase K241A mutant in complex with D-ACL (external aldimine) | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class-III, [6-methyl-5-oxidanyl-4-[(~{E})-[(3~{R})-2-oxidanylideneazepan-3-yl]iminomethyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
1EG6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF D(CG(5-BRU)ACG) COMPLEXES TO A PHENAZINE | | Descriptor: | 5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3', 9-BROMO-PHENAZINE-1-CARBOXYLIC ACID (2-DIMETHYLAMINO-ETHYL)-AMIDE, BROMIDE ION, ... | | Authors: | Cardin, C.J, Denny, W.A, Hobbs, J.R, Thorpe, J.H. | | Deposit date: | 2000-02-14 | | Release date: | 2001-01-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Guanine specific binding at a DNA junction formed by d[CG(5-BrU)ACG](2) with a topoisomerase poison in the presence of Co(2+) ions.
Biochemistry, 39, 2000
|
|
1EGA
 
 | | CRYSTAL STRUCTURE OF A WIDELY CONSERVED GTPASE ERA | | Descriptor: | PROTEIN (GTP-BINDING PROTEIN ERA), SULFATE ION | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1998-12-01 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5M4R
 
 | | Structural tuning of CD81LEL (space group C2) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, SULFATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
9GET
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)pent-1-en-1-yl)-4-methyl-8-(morpholine-4-carbonyl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one | | Descriptor: | (4~{R})-6-[(~{E})-5-(7-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)pent-1-enyl]-4-methyl-8-morpholin-4-ylcarbonyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Monoglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)pent-1-en-1-yl)-4-methyl-8-(morpholine-4-carbonyl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one
To Be Published
|
|
5M5A
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, K-252A, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-10-21 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
1EI1
 
 | | DIMERIZATION OF E. COLI DNA GYRASE B PROVIDES A STRUCTURAL MECHANISM FOR ACTIVATING THE ATPASE CATALYTIC CENTER | | Descriptor: | DNA GYRASE B, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Brino, L, Urzhumtsev, A, Oudet, P, Moras, D. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center.
J.Biol.Chem., 275, 2000
|
|
5M5F
 
 | | Thermolysin in complex with inhibitor and krypton | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-21 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
1ERZ
 
 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | Descriptor: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | Authors: | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | Deposit date: | 2000-04-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
