7BYN
 
 | | Cryo-EM structure of human KCNQ4 with linopirdine | | Descriptor: | 1-phenyl-3,3-bis(pyridin-4-ylmethyl)indol-2-one, Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7BYL
 
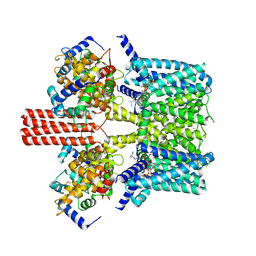 | | Cryo-EM structure of human KCNQ4 | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
5GGM
 
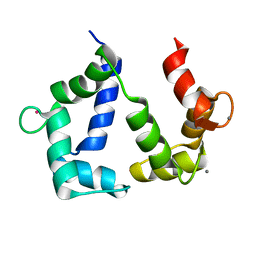 | | The NMR structure of calmodulin in CTAB reverse micelles | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3V7X
 
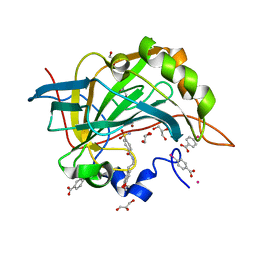 | | Complex of human carbonic anhydrase II with N-[2-(3,4-dimethoxyphenyl)ethyl]-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, MERCURIBENZOIC ACID, ... | | Authors: | Mader, P, Brynda, J, Rezacova, P. | | Deposit date: | 2011-12-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
7BFA
 
 | |
7BG5
 
 | |
5AMG
 
 | |
3VHX
 
 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
7BHH
 
 | |
5AML
 
 | |
5HIT
 
 | |
3VBD
 
 | | Complex of human carbonic anhydrase II with 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide | | Descriptor: | 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Mader, P, Brynda, J, Rezacova, P. | | Deposit date: | 2012-01-02 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
3ECV
 
 | |
7CR7
 
 | | human KCNQ2-CaM in complex with retigabine | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
3ECU
 
 | |
3EFI
 
 | | Carbonic anhydrase activators: Kinetic and X-ray crystallographic study for the interaction of d- and l-tryptophan with the mammalian isoforms I-XIV | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, TRYPTOPHAN, ... | | Authors: | Temperini, C, Innocenti, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-09-09 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbonic anhydrase activators: kinetic and X-ray crystallographic study for the interaction of D- and L-tryptophan with the mammalian isoforms I-XIV
Bioorg.Med.Chem., 16, 2008
|
|
7CR3
 
 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CA2
 
 | |
7CR4
 
 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
5BRW
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BNL
 
 | | Deciphering the Mechanism of Carbonic Anhydrase Inhibition with Coumarins and Thiocoumarins | | Descriptor: | (2E)-3-(2-HYDROXYPHENYL)ACRYLIC ACID, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Maresca, A, Pochet, L, Masereel, B, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2015-05-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the mechanism of carbonic anhydrase inhibition with coumarins and thiocoumarins.
J.Med.Chem., 53, 2010
|
|
5C0N
 
 | |
3EWT
 
 | | Crystal Structure of calmodulin complexed with a peptide | | Descriptor: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | Deposit date: | 2008-10-16 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
