5M5B
 
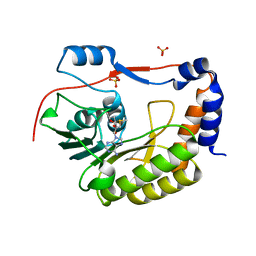 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
1EVW
 
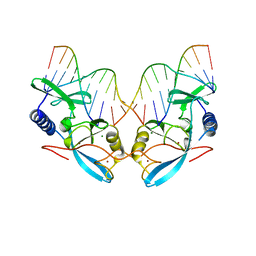 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
5LT6
 
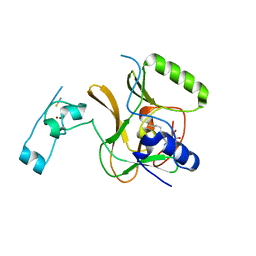 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
5LTJ
 
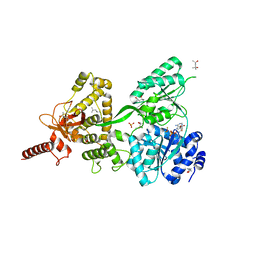 | | Crystal structure of the Prp43-ADP-BeF3 complex (in orthorhombic space group) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tauchert, M.J, Ficner, R. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the mechanism of the DEAH-box RNA helicase Prp43.
Elife, 6, 2017
|
|
1EWP
 
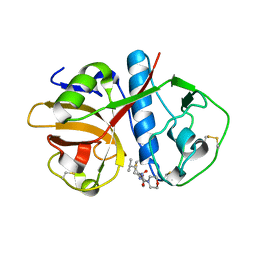 | | CRUZAIN BOUND TO MOR-LEU-HPQ | | Descriptor: | CRUZAIN, N-[(3S)-1-fluoro-2-oxo-5-phenylpentan-3-yl]-N~2~-(morpholin-4-ylcarbonyl)-L-leucinamide | | Authors: | Gillmor, S.A. | | Deposit date: | 2000-04-26 | | Release date: | 2000-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Chapter 3: X-ray Structures of Complexes of Cruzain with Designed Covalent Inhibitors
Enzyme-ligand Interactions, Inhibition and Specificity, 1998
|
|
1EWY
 
 | | ANABAENA PCC7119 FERREDOXIN:FERREDOXIN-NADP+-REDUCTASE COMPLEX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I, FERREDOXIN-NADP REDUCTASE, ... | | Authors: | Morales, R, Charon, M.H, Frey, M. | | Deposit date: | 2000-04-28 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic studies of the interaction between the ferredoxin-NADP+ reductase and ferredoxin from the cyanobacterium Anabaena: looking for the elusive ferredoxin molecule.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EXD
 
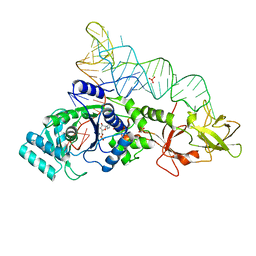 | | CRYSTAL STRUCTURE OF A TIGHT-BINDING GLUTAMINE TRNA BOUND TO GLUTAMINE AMINOACYL TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMINE TRNA APTAMER, GLUTAMINYL-TRNA SYNTHETASE, ... | | Authors: | Bullock, T.L, Sherlin, L.D, Perona, J.J. | | Deposit date: | 2000-05-02 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary core rearrangements in a tight binding transfer RNA aptamer.
Nat.Struct.Biol., 7, 2000
|
|
9G7B
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and loreclezole | | Descriptor: | 1-[(~{Z})-2-chloranyl-2-(2,4-dichlorophenyl)ethenyl]-1,2,4-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-07-20 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | CryoEM structure of human rho1 GABAA receptor in complex with neurosteroid
To be published
|
|
5M5K
 
 | | S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine and cordycepin | | Descriptor: | 3'-DEOXYADENOSINE, ACETATE ION, ADENOSINE, ... | | Authors: | Manszewski, T, Mueller-Dieckamann, J, Jaskolski, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
5LGA
 
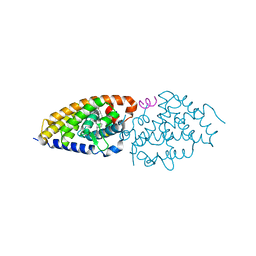 | | Structural analysis and biological activities of BXL0124, a Gemini analog of Vitamin D | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(6~{R})-1,1,1-tris(fluoranyl)-10-methyl-2,10-bis(oxidanyl)-2-(trifluoromethyl)undeca-3,8-diyn-6-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, SRC-2, Vitamin D3 receptor A | | Authors: | Belorusova, A.Y, Rochel, N. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and biological activities of BXL0124, a gemini analog of vitamin D.
J. Steroid Biochem. Mol. Biol., 173, 2017
|
|
1EY6
 
 | |
9G6O
 
 | | Xylose Isomerase collected at 45C using time-resolved serial synchrotron crystallography with Glucose at 60 seconds | | Descriptor: | D-glucose, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Schulz, E.C, Prester, A, Stetten, D.V, Gore, G, Hatton, C.E, Bartels, K, Leimkohl, J.P, Schikora, H, Ginn, H.M, Tellkamp, F, Mehrabi, P. | | Deposit date: | 2024-07-18 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
9G6N
 
 | | Xylose Isomerase collected at 40C using time-resolved serial synchrotron crystallography with Glucose at 60 seconds | | Descriptor: | D-glucose, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Schulz, E.C, Prester, A, Stetten, D.V, Gore, G, Hatton, C.E, Bartels, K, Leimkohl, J.P, Schikora, H, Ginn, H.M, Tellkamp, F, Mehrabi, P. | | Deposit date: | 2024-07-18 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
1EYD
 
 | |
1EYL
 
 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
5LUL
 
 | | Structure of a triple variant of cutinase 2 from Thermobifida cellulosilytica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase 2 | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
1EYR
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | Descriptor: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE, CYTIDINE-5'-DIPHOSPHATE | | Authors: | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | Deposit date: | 2000-05-08 | | Release date: | 2001-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
5LUR
 
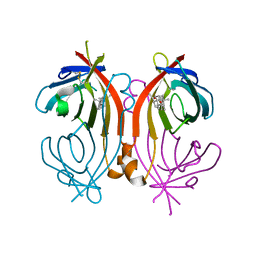 | | An avidin mutant | | Descriptor: | Avidin, CHLORIDE ION, PROGESTERONE | | Authors: | Agrawal, N, Lehtonen, S.I, Riihimaki, T.A, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An avidin mutant
TO BE PUBLISHED
|
|
1E95
 
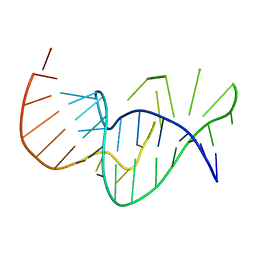 | | Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting | | Descriptor: | RNA (5'-(*GP*CP*GP*GP*CP*CP*AP*GP*CP*UP*CP* CP*AP*GP*GP*CP*CP*GP*CP*CP*AP*AP*AP*CP* AP*AP*UP*AP*UP*GP*GP*AP*GP*CP*AP*C)-3') | | Authors: | Michiels, P.J.A, Versleyen, A, Pleij, C.W.A, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2000-10-09 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pseudoknot of Srv-1 RNA, Involved in Ribosomal Frameshifting
J.Mol.Biol., 310, 2001
|
|
5LUZ
 
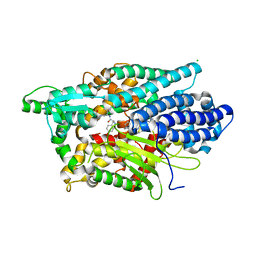 | | Structure of Human Neurolysin (E475Q) in complex with neurotensin peptide products | | Descriptor: | CHLORIDE ION, GLYCEROL, Neurolysin, ... | | Authors: | Masuyer, G, Berntsson, R.P.-A, Teixeira, P.F, Kmiec, B, Glaser, E, Stenmark, P. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Peptide Binding and Cleavage by the Human Mitochondrial Peptidase Neurolysin.
J. Mol. Biol., 430, 2018
|
|
1E6A
 
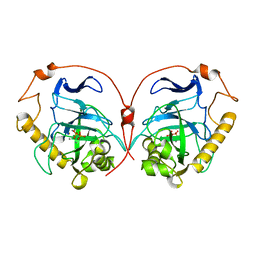 | | Fluoride-inhibited substrate complex of Saccharomyces cerevisiae inorganic pyrophosphatase | | Descriptor: | FLUORIDE ION, INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-08-09 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward a quantum-mechanical description of metal-assisted phosphoryl transfer in pyrophosphatase.
Proc. Natl. Acad. Sci. U.S.A., 98, 2001
|
|
9GA1
 
 | |
1E4Y
 
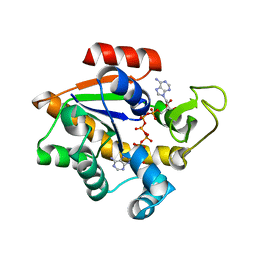 | |
5LVQ
 
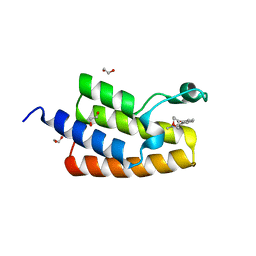 | | Crystal structure of human PCAF bromodomain in complex with compound-D (CPD-D), N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
1EM0
 
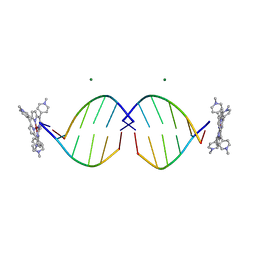 | | COMPLEX OF D(CCTAGG) WITH TETRA-[N-METHYL-PYRIDYL] PORPHYRIN | | Descriptor: | DNA (5'-D(*(CBR)P*CP*TP*AP*GP*G)-3'), MAGNESIUM ION, TETRA[N-METHYL-PYRIDYL] PORPHYRIN-NICKEL | | Authors: | Neidle, S, Sanderson, M, Bennett, M, Krah, A, Wien, F, Garman, E, McKenna, R. | | Deposit date: | 2000-03-14 | | Release date: | 2000-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
