1EP3
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B. DATA COLLECTED UNDER CRYOGENIC CONDITIONS. | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
5LSU
 
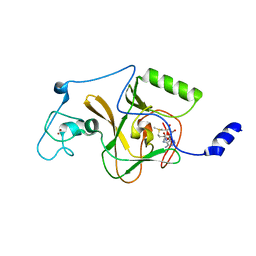 | |
1EQ1
 
 | |
5LNB
 
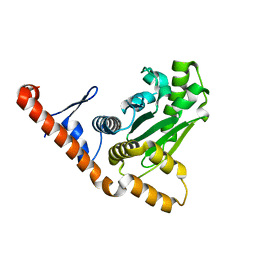 | |
1ES2
 
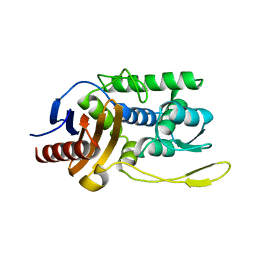 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES9
 
 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
5LNF
 
 | | Solution NMR structure of farnesylated PEX19, C-terminal domain | | Descriptor: | FARNESYL, Peroxisomal biogenesis factor 19 | | Authors: | Emmanouilidis, L, Schuetz, U, Tripsianes, K, Madl, T, Radke, J, Rucktaeschel, R, Wilmanns, M, Schliebs, W, Erdmann, R, Sattler, M. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19.
Nat Commun, 8, 2017
|
|
1ESK
 
 | | SOLUTION STRUCTURE OF NCP7 FROM HIV-1 | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Morellet, N, Demene, H, Teilleux, V, Huynh-Dinh, T, de Rocquigny, H, Fournie-Zaluski, M.-C, Roques, B.P. | | Deposit date: | 2000-04-10 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of (12-53)NCp7 of HIV-1
To be Published
|
|
9FZT
 
 | |
5LTK
 
 | |
1ETN
 
 | |
1ETX
 
 | |
1E7I
 
 | |
5LNP
 
 | |
9G5Q
 
 | |
7RUJ
 
 | | E. coli cysteine desulfurase SufS N99A | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
5LNY
 
 | | HSP90 WITH indazole derivative | | Descriptor: | 6-Hydroxy-3-(piperidine-1-carbonyl)-1H-indazole-5-carboxylic acid methyl-(4-morpholin-4-yl-phenyl)-amide, Heat shock protein HSP 90-alpha | | Authors: | Graedler, U, Amaral, M, Schuetz, D. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
9G78
 
 | |
1E5B
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1E5E
 
 | | METHIONINE GAMMA-LYASE (MGL) FROM TRICHOMONAS VAGINALIS IN COMPLEX WITH PROPARGYLGLYCINE | | Descriptor: | GLYCEROL, METHIONINE GAMMA-LYASE, N-(HYDROXY{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)NORVALINE, ... | | Authors: | Goodall, G, Mottram, J.C, Coombs, G.H, Lapthorn, A.J. | | Deposit date: | 2000-07-25 | | Release date: | 2001-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structure and Proposed Catalytic Mechanism of Methionine Gamma-Lyase
To be Published
|
|
5LUK
 
 | | Structure of a double variant of cutinase 2 from Thermobifida cellulosilytica | | Descriptor: | CHLORIDE ION, Cutinase 2, MAGNESIUM ION | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
1E5Z
 
 | |
5LUQ
 
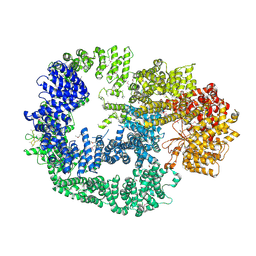 | | Crystal Structure of Human DNA-dependent Protein Kinase Catalytic Subunit (DNA-PKcs) | | Descriptor: | C-terminal fragment of KU80 (KU80ct194), DNA-dependent protein kinase catalytic subunit,DNA-dependent Protein Kinase Catalytic Subunit,DNA-dependent protein kinase catalytic subunit | | Authors: | Sibanda, B.L, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair.
Science, 355, 2017
|
|
5LOR
 
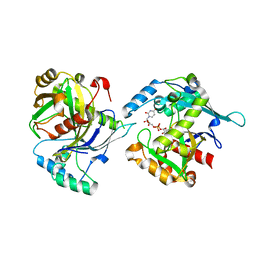 | | human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2016-08-09 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structural and functional studies of human NUDT22
To Be Published
|
|
1E7W
 
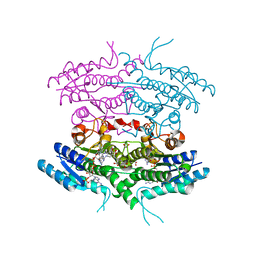 | | One active site, two modes of reduction correlate the mechanism of leishmania pteridine reductase with pterin metabolism and antifolate drug resistance in trpanosomes | | Descriptor: | 1,2-ETHANEDIOL, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gourley, D.G, Hunter, W.N. | | Deposit date: | 2000-09-11 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pteridine Reductase Mechanism Correlates Pterin Metabolism with Drug Resistance in Trypanosomatid Parasites.
Nat.Struct.Biol., 8, 2001
|
|
