4OOX
 
 | |
1CAD
 
 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
1UCY
 
 | |
1CAA
 
 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
3UMJ
 
 | | Crystal Structure of D311E Lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ruslan, R, Rahman, R.N.Z.R.A, Leow, T.C, Ali, M.S.M, Basri, M, Salleh, A.B. | | Deposit date: | 2011-11-13 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Thermal Stability via Outer-Loop Ion Pair Interaction of Mutated T1 Lipase from Geobacillus zalihae Strain T1
Int J Mol Sci, 13, 2012
|
|
7EPC
 
 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
1UHE
 
 | | Crystal structure of aspartate decarboxylase, isoaspargine complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, N~2~-(2-AMINO-1-METHYL-2-OXOETHYLIDENE)ASPARAGINATE | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
7EPF
 
 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
4EKT
 
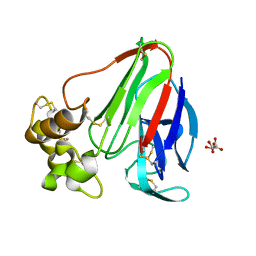 | | Final Thaumatin Structure for Radiation Damage Experiment at 180 K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-09 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EL3
 
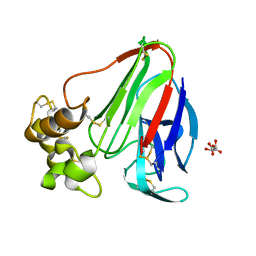 | | Final Thaumatin Structure for Radiation Damage Experiment at 240 K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2C29
 
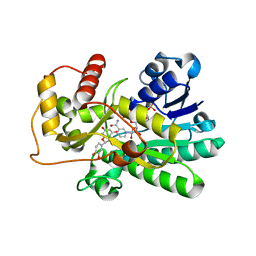 | | Structure of dihydroflavonol reductase from Vitis vinifera at 1.8 A. | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, DIHYDROFLAVONOL 4-REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Granier, T, D'Estaintot, B.L, Hamdi, S, Gallois, B. | | Deposit date: | 2005-09-27 | | Release date: | 2006-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis.
J.Mol.Biol., 368, 2007
|
|
4EPE
 
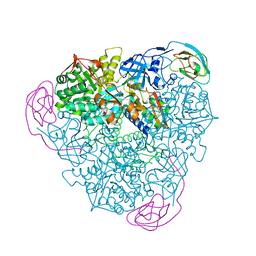 | | Final Urease Structure for Radiation Damage Experiment at 300 K | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EL7
 
 | | Initial Thaumatin Structure for Radiation Damage Experiment at 300 K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1YTB
 
 | | CRYSTAL STRUCTURE OF A YEAST TBP/TATA-BOX COMPLEX | | Descriptor: | DNA (29MER), PROTEIN (TATA BINDING PROTEIN (TBP)) | | Authors: | Kim, Y, Geiger, J.H, Hahn, S, Sigler, P.B. | | Deposit date: | 1994-09-28 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a yeast TBP/TATA-box complex.
Nature, 365, 1993
|
|
2JHM
 
 | | Structure of globular heads of M-ficolin at neutral pH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, FICOLIN-1, ... | | Authors: | Garlatti, V, Martin, L, Gout, E, Reiser, J.B, Arlaud, G.J, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Basis for Innate Immune Sensing by M-Ficolin and its Control by a Ph-Dependent Conformational Switch.
J.Biol.Chem., 282, 2007
|
|
5O83
 
 | | Discovery of CDZ173 (leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors | | Descriptor: | Leniolisib, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of CDZ173 (Leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
3E3Z
 
 | | Crystal structure of bovine coupling Factor B bound with phenylarsine oxide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP synthase subunit s, mitochondrial, ... | | Authors: | Stroud, R.M, Lee, J.K, Belogrudov, G.I. | | Deposit date: | 2008-08-08 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of bovine mitochondrial factor B at 0.96-A resolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3E4G
 
 | | Crystal structure of bovine coupling Factor B, G28E mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP synthase subunit s, mitochondrial, ... | | Authors: | Stroud, R.M, Lee, J.K, Belogrudov, G.I. | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal structure of bovine mitochondrial factor B at 0.96-A resolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3OF1
 
 | | Crystal Structure of Bcy1, the Yeast Regulatory Subunit of PKA | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase regulatory subunit | | Authors: | Rinaldi, J, Wu, J, Yang, J, Ralston, C.Y, Sankaran, B, Moreno, S, Taylor, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of Yeast Regulatory Subunit: A Glimpse into the Evolution of PKA Signaling.
Structure, 18, 2010
|
|
3ZM7
 
 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPCP | | Descriptor: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
2WLC
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
3OHR
 
 | | Crystal structure of fructokinase from bacillus subtilis complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative fructokinase, SULFATE ION, ... | | Authors: | Nocek, B, Volkart, L, Cuff, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
4GOW
 
 | | Crystal Structure of Ca2+/CaM:Kv7.4 (KCNQ4) B helix complex | | Descriptor: | CALCIUM ION, Calmodulin, Potassium voltage-gated channel subfamily KQT member 4 | | Authors: | Xu, Q, Chang, A, Tolia, A, Minor Jr, D.L. | | Deposit date: | 2012-08-20 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Ca(2+)/CaM:Kv7.4 (KCNQ4) B-helix complex provides insight into M current modulation.
J.Mol.Biol., 425, 2013
|
|
1I8A
 
 | | FAMILY 9 CARBOHYDRATE-BINDING MODULE FROM THERMOTOGA MARITIMA XYLANASE 10A WITH GLUCOSE | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Warren, R.A.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-03-12 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Biochemistry, 40, 2001
|
|
