5OTT
 
 | |
1U9E
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-397 | | Descriptor: | 2-(4-HYDROXY-PHENYL)BENZOFURAN-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design Of Estrogen Receptor-beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
8HBA
 
 | |
8HB1
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8HB8
 
 | |
8HB3
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
4WVV
 
 | |
2PEV
 
 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution exceeds concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-03 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
8H4V
 
 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
8HB5
 
 | | Crystal structure of Mincle in complex with HD-275 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
6S53
 
 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | Authors: | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
2QPQ
 
 | | Structure of Bug27 from Bordetella pertussis | | Descriptor: | CITRIC ACID, protein Bug27 | | Authors: | Herrou, J, Bompard, C. | | Deposit date: | 2007-07-25 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-based mechanism of ligand binding for periplasmic solute-binding protein of the Bug family.
J.Mol.Biol., 373, 2007
|
|
5HM4
 
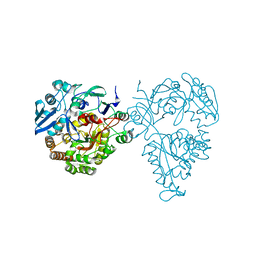 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | Descriptor: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | Authors: | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
6V95
 
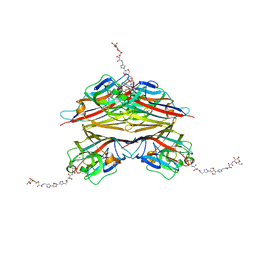 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-tartaramidoyl derivative (diNGT) | | Descriptor: | (2R,3R)-N-[(1-{(3S,3aR,6S,6aR)-6-[4-({[(2R,3R)-2,3-dihydroxy-4-oxo-4-{[(2R,3R,4R,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]amino}butanoyl]amino}methyl)-1H-1,2,3-triazol-1-yl]hexahydrofuro[3,2-b]furan-3-yl}-1H-1,2,3-triazol-4-yl)methyl]-2,3-dihydroxy-N'-[(2R,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]butanediamide (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5DHG
 
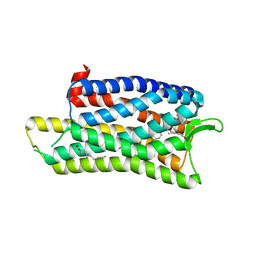 | | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with C-35 (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-benzyl-N-{3-[4-(2,6-dichlorophenyl)piperidin-1-yl]propyl}-D-prolinamide, OLEIC ACID, ... | | Authors: | Miller, R.L, Thompson, A.A, Trapella, C, Guerrini, R, Malfacini, D, Patel, N, Han, G.W, Cherezov, V, Calo, G, Katritch, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-08-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor.
Structure, 23, 2015
|
|
6MNI
 
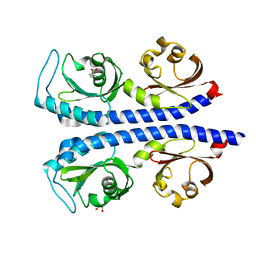 | | Structure of the tandem CACHE domain of PscC | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Johnston, J.M, Gerth, M.L, Ehrhardt, M.K.G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure of a double CACHE chemoreceptor ligand-binding domain from Pseudomonas syringae provides insights into the basis of proline recognition.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
6VGF
 
 | | Peanut lectin complexed with divalent S-beta-D-thiogalactopyranosyl beta-D-glucopyranoside derivative (diSTGD) | | Descriptor: | (2S,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({4-[({[(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-2-yl]methyl}sulfanyl)methyl]-1H-1,2,3-triazol-1-yl}methyl)tetrahydro-2H-pyran-2-yl]oxy}tetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4WVW
 
 | | Chicken Galectin-8 N-terminal domain complexed with 3'-sialyl-lactose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galectin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2014-11-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combining Crystallography and Hydrogen-Deuterium Exchange to Study Galectin-Ligand Complexes.
Chemistry, 21, 2015
|
|
5OBE
 
 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-MBI), MBI=methyl-benzimidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, pentakis(oxidaniumyl)-(oxidaniumylidynemethyl)ruthenium, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
8I3Z
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN at 1.67 angstrom | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3'), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
3UWW
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with 3-phosphoglyceric acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-PHOSPHOGLYCERIC ACID, SODIUM ION, ... | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
5OBD
 
 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, tris(oxidaniumylidynemethyl)-tris(oxidanyl)ruthenium | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
3UWY
 
 | |
4TT4
 
 | | Crystal structure of ATAD2A bromodomain complexed with H3(1-21)K14Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, Histone H3(1-21)K4Ac, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
7R8P
 
 | |
