7X2D
 
 | | Cryo-EM structure of the tavapadon-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 1,5-dimethyl-6-[2-methyl-4-[3-(trifluoromethyl)pyridin-2-yl]oxy-phenyl]pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
4C6M
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to the inhibitor fluoroorotate at pH 7.0 | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
5BVW
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
9IZE
 
 | | Acarbose hydrolase from human gut microbiota K. grimontii TD1, Apg mutant enzyme D336A, complexed with acarviosine-glucose | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-5-[[(1~{S},4~{R},5~{S},6~{S})-3-(hydroxymethyl)-4,5,6-tris(oxidanyl)cyclohex-2-en-1-yl]amino]-6-methyl-oxane-2,3,4-triol-(1-4)-alpha-D-glucopyranose, Maltodextrin glucosidase | | Authors: | Zhou, J.H, Huang, J.Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Molecular insights of acarbose metabolization catalyzed by acarbose-preferred glucosidase.
Nat Commun, 16, 2025
|
|
5STJ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P02H11 from the F2X-Universal Library | | Descriptor: | 1-(3-methoxy-4-methylphenyl)methanamine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
3LHH
 
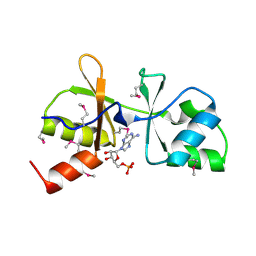 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
7I6P
 
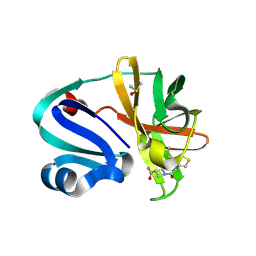 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037779-001 (A71EV2A-x4998) | | Descriptor: | 1-{[4-(methylsulfanyl)thiophen-3-yl]acetyl}azetidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
5C9P
 
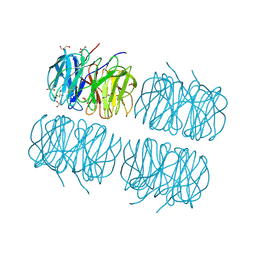 | | Crystal structure of recombinant PLL lectin complexed with L-fucose from Photorhabdus luminescens at 1.75 A resolution | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
5STI
 
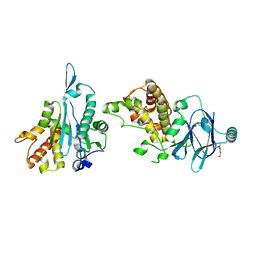 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P02H11 from the F2X-Universal Library | | Descriptor: | 1-(3-methoxy-4-methylphenyl)methanamine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FHM
 
 | |
4OYD
 
 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | Authors: | Shen, B, Procko, E, Baker, D, Stoddard, B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
5C2H
 
 | | PDE10 complexed with 6-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]-5-methyl-2-[3-(2-quinolyl)propoxy]pyrimidin-4-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 6-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]-5-methyl-2-[3-(quinolin-2-yl)propoxy]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Optimization of a Series of Pyrimidine-Based Phosphodiesterase 10A (PDE10A) Inhibitors through Fragment Screening, Structure-Based Design, and Parallel Synthesis.
J.Med.Chem., 58, 2015
|
|
7ZTN
 
 | | Crystal structure of fungal CE16 acetyl xylan esterase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Kosinas, C, Pentari, C, Zerva, A, Topakas, E. | | Deposit date: | 2022-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The role of CE16 exo-deacetylases in hemicellulolytic enzyme mixtures revealed by the biochemical and structural study of the novel TtCE16B esterase.
Carbohydr Polym, 327, 2024
|
|
9F0M
 
 | |
5GGS
 
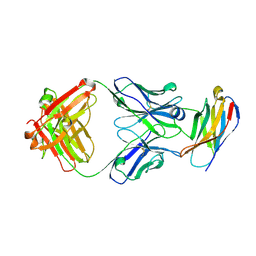 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5M63
 
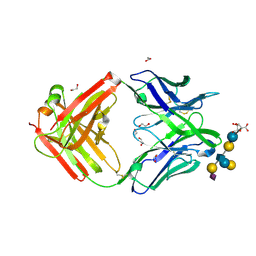 | | Crystal structure of group B Streptococcus type III DP2 oligosaccharide bound to Fab NVS-1-19-5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-[bis(oxidanyl)methyl]-5-(hydroxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, H chain of Fab NVS-1-19-5, ... | | Authors: | Carboni, F, Adamo, R, Veggi, D, Rappuoli, R, Malito, E, Margarit, I.R, Berti, F. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GID
 
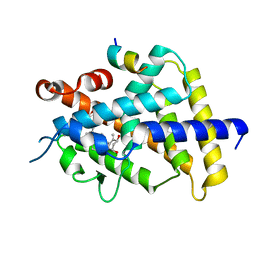 | | Crystal structure of VDR in complex with DLAM-4 (C2 form) | | Descriptor: | (3R,5S)-5-[(2R)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[(2Z)-2-[(3S,5R)-2-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]propyl]-3-methyl-3-oxidanyl-1-(4-phenylbutyl)pyrrolidin-2-one, SRC1, Vitamin D3 receptor | | Authors: | Asano, L, Shimizu, T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Regulation of the vitamin D receptor by vitamin D lactam derivatives.
Febs Lett., 590, 2016
|
|
6DZ3
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-(((3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl)thio)methyl)pyrrolidin-3-ol | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl]sulfanyl}methyl)pyrrolidin-3-ol, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
7T1D
 
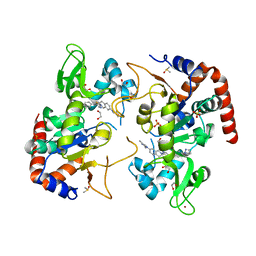 | | Human SIRT2 in complex with small molecule 359 | | Descriptor: | 1,2-ETHANEDIOL, 7-(2,4-dimethyl-1H-imidazol-1-yl)-2-(5-{[4-(1H-pyrazol-1-yl)phenyl]methyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Kulp, J.L, Remiszewski, S, Todd, M, Chiang, L.W. | | Deposit date: | 2021-12-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric inhibitor of sirtuin 2 deacetylase activity exhibits broad-spectrum antiviral activity.
J.Clin.Invest., 133, 2023
|
|
5CGP
 
 | |
5GTO
 
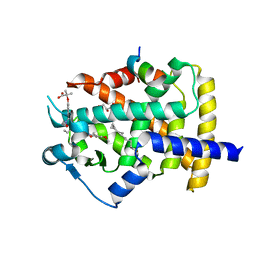 | | Human PPARgamma ligand binding dmain complexed with S35 | | Descriptor: | 2-[4-[5-[(1~{S})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
6TNZ
 
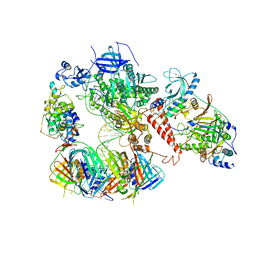 | | Human polymerase delta-FEN1-PCNA toolbelt | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
8DGY
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d (high resolution) | | Descriptor: | 3C-like proteinase, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
9CTB
 
 | | Tri-complex of zoldonrasib (RMC-9805), KRAS G12D, and CypA | | Descriptor: | (2R)-2-cyclopentyl-2-{(5S)-7-[(2R)-3-cyclopropyl-2-(methylamino)propanoyl]-2,7-diazaspiro[4.4]nonan-2-yl}-N-[(2S,6R,8S,10R,14S,21M)-21-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-22-(2,2,2-trifluoroethyl)-5,16-dioxa-2,10,22,28-tetraazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),20,23,26-tetraen-8-yl]acetamide (non-preferred name), CHLORIDE ION, Isoform 2B of GTPase KRas, ... | | Authors: | Zhang, D, Bar Ziv, T, Tomlinson, A.C.A, Saldajeno-Concar, M, Yano, J.K, Knox, J.E. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Science, 389, 2025
|
|
