5AXB
 
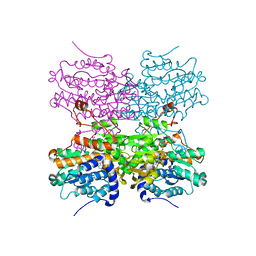 | | Crystal structure of mouse SAHH complexed with noraristeromycin | | Descriptor: | (1S,2R,3S,4R)-4-(6-aminopurin-9-yl)cyclopentane-1,2,3-triol, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse SAHH complexed with noraristeromycin
To Be Published
|
|
6TO6
 
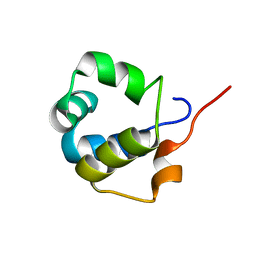 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4OJU
 
 | |
6MDC
 
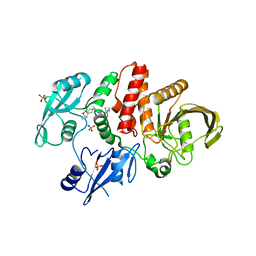 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor Pyrazolo-pyrimidinone 1 SHP389 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-[3-chloro-2-(cyclopropylamino)pyridin-4-yl]-5-methyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Fodor, M, Stams, T. | | Deposit date: | 2018-09-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of Fused Bicyclic Allosteric SHP2 Inhibitors.
J. Med. Chem., 62, 2019
|
|
7UCW
 
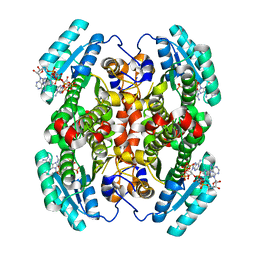 | | Structure of mouse Decr1 in complex with 2'-5' oligoadenylate | | Descriptor: | Decr1 protein, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Govande, A.A, Kranzusch, P.J. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RNase L-activating 2'-5' oligoadenylates bind ABCF1, ABCF3 and Decr-1.
J.Gen.Virol., 104, 2023
|
|
5D5C
 
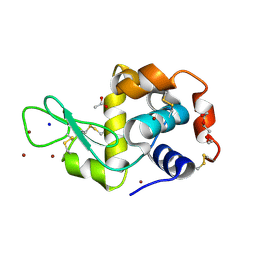 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2AMY
 
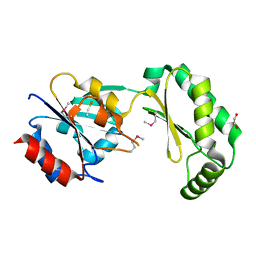 | | X-Ray Structure of Human Phosphomannomutase 2 (PMM2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, Phosphomannomutase 2 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-10 | | Release date: | 2005-08-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-Ray Structure of Human Phosphomannomutase 2 (PMM2)
To be Published
|
|
5TRI
 
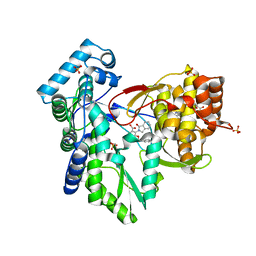 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid, NS5B RNA-DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and initial optimization of alkoxyanthranilic acid derivatives as inhibitors of HCV NS5B polymerase.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7B53
 
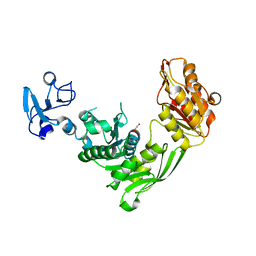 | | Crystal structure of MurE from E.coli | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5D7P
 
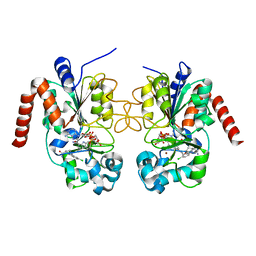 | | Crystal structure of human Sirt2 in complex with ADPR and EX-243 | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6TLW
 
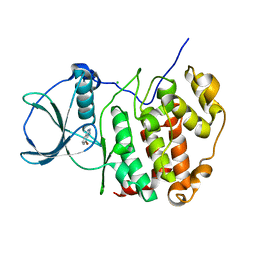 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4-BROMOBENZOTRIAZOLE | | Descriptor: | 7-bromanyl-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
4ZGF
 
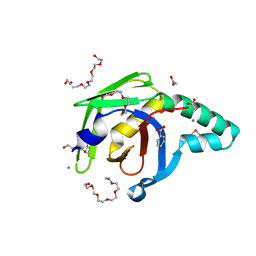 | |
4YZ3
 
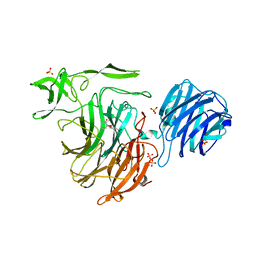 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Streptococcus pneumoniae NanC.
To Be Published
|
|
4PE4
 
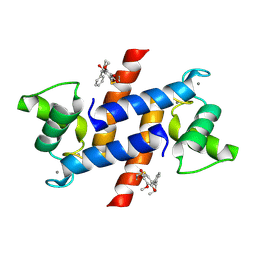 | | Crystal Structure of Calcium-loaded S100B bound to SC1475 | | Descriptor: | 2,3-dimethoxy-5-[(1S)-1-phenylpropyl]benzene-1,4-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.178 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
4Z16
 
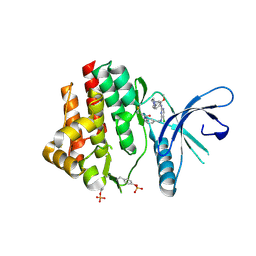 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | Descriptor: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | Authors: | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5DGH
 
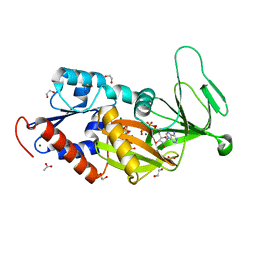 | |
5YQ8
 
 | |
5DH9
 
 | |
5EKA
 
 | | HU DNA-binding protein from Thermus thermophilus | | Descriptor: | DNA-binding protein HU, GLYCEROL | | Authors: | Papageorgiou, A, Adam, P, Stavros, P, Nounesis, G, Meijers, R, Petratos, K, Vorgias, C.E. | | Deposit date: | 2015-11-03 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses.
Extremophiles, 20, 2016
|
|
6LYK
 
 | | Crystal Structure of R1263A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
4Z6H
 
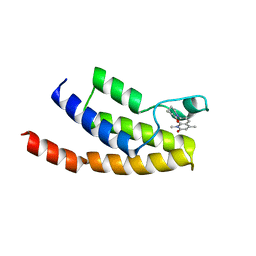 | | Crystal structure of BRD9 bromodomain in complex with a valerolactam quinolone ligand | | Descriptor: | 1,4-dimethyl-7-(2-oxopiperidin-1-yl)quinolin-2(1H)-one, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DJL
 
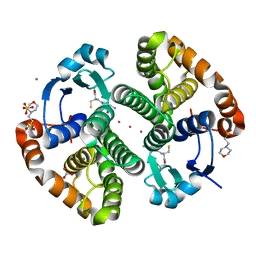 | |
5WZX
 
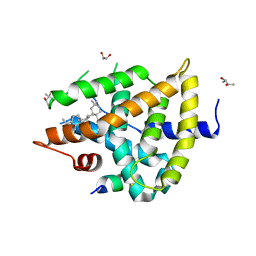 | | Structural basis for a pentacyclic oleanane-type triterpenoid as a ligand of FXR | | Descriptor: | (4aR,6aR,6aS,6bS,8aS,9R,12aR,14bR)-2,2,6a,6b,9,12a-hexamethyl-10-oxidanylidene-1,3,4,5,6,6a,7,8,8a,9,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid, (R,R)-2,3-BUTANEDIOL, Bile acid receptor, ... | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of an Oleanane-Type Triterpene Hedragonic Acid as a Novel Farnesoid X Receptor Ligand with Liver Protective Effects and Anti-inflammatory Activity
Mol. Pharmacol., 93, 2018
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
