5SH6
 
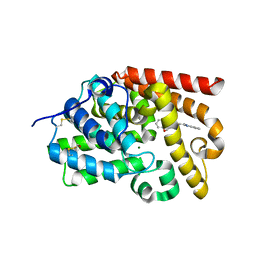 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c12c(cccc1)nc(n2C)CCN4CCOc3ccccc3C4=O, micromolar IC50=2.844764 | | Descriptor: | 4-[2-(1-methyl-1H-benzimidazol-2-yl)ethyl]-3,4-dihydro-1,4-benzoxazepin-5(2H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3LL7
 
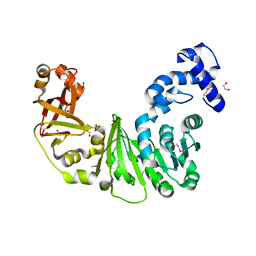 | | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase | | Authors: | Chang, C, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83
To be Published
|
|
7SYR
 
 | | Structure of the wt IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(wt). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
5SI8
 
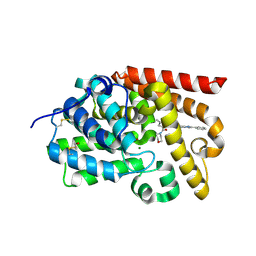 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cn(nc1C(=O)Nc3cc2nc(cn2cc3C#N)c4ccccc4)C)C(=O)N5CCOCC5, micromolar IC50=0.060125 | | Descriptor: | MAGNESIUM ION, N-[(4R)-6-cyano-2-phenylimidazo[1,2-a]pyridin-7-yl]-1-methyl-4-(morpholine-4-carbonyl)-1H-pyrazole-3-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5FPM
 
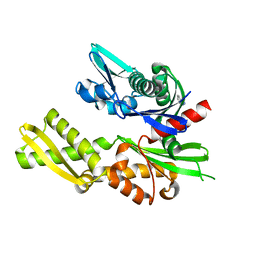 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 5-phenyl-1,3,4-oxadiazole-2-thiol (AT809) in an alternate binding site. | | Descriptor: | 5-PHENYL-1,3,4-OXADIAZOLE-2-THIOL, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3OVB
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
7AHF
 
 | |
9CT4
 
 | | HsSTING with diABZI and C53, curved conformation | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-1,3-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-1,3-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Gharpure, A, Sulpizio, A, Lairson, L.L, Ward, A.B. | | Deposit date: | 2024-07-24 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Distinct oligomeric assemblies of STING induced by non-nucleotide agonists.
Nat Commun, 16, 2025
|
|
5SI9
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c12nc(cn1ccc(n2)NC(c3c(cnn3C)C(NCCO)=O)=O)c4ccccc4, micromolar IC50=0.054287 | | Descriptor: | MAGNESIUM ION, N~4~-(2-hydroxyethyl)-1-methyl-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7I1Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with NegAcid1-Am02 | | Descriptor: | DIMETHYL SULFOXIDE, Serine protease NS3, Serine protease subunit NS2B, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Balaji, G, Phelps, J, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2VRF
 
 | | CRYSTAL STRUCTURE OF THE HUMAN BETA-2-SYNTROPHIN PDZ DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, BETA-2-SYNTROPHIN | | Authors: | Sun, Z, Roos, A.K, Pike, A.C.W, Pilka, E.S, Cooper, C, Elkins, J.M, Murray, J, Arrowsmith, C.H, Doyle, D, Edwards, A, von Delft, F, Bountra, C, Oppermann, U. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Beta-2-Syntrophin Pdz Domain
To be Published
|
|
5SK0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N2(c1ccc(cn1)Cl)CC[C@H](C2)NC(c3nn(cc3c4ccncc4)C)=O, micromolar IC50=0.708457 | | Descriptor: | MAGNESIUM ION, N-[(3R)-1-(5-chloropyridin-2-yl)pyrrolidin-3-yl]-1-methyl-4-(pyridin-4-yl)-1H-pyrazole-3-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5ZEP
 
 | | M. smegmatis hibernating state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5SJ9
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1(C)c(nc(c1)c2ccccc2)CCNC(c3cccn4c(nnc34)C(C)C)=O, micromolar IC50=0.024844 | | Descriptor: | (4S)-N-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]-3-(propan-2-yl)[1,2,4]triazolo[4,3-a]pyridine-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7AHR
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
5SE3
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(cc1nc(nn1cc2)c3ccccc3)NC(c4ccnc(c4)Cl)=O, micromolar IC50=0.045025 | | Descriptor: | 2-chloro-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]pyridine-4-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEG
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(c(C(=O)NC1COC1)cc3c(c2)nc([nH]3)c4ccccc4)NC(C5=CC(=CN(C5=O)C)Br)=O, micromolar IC50=0.006706 | | Descriptor: | 5-[(5-bromo-1-methyl-2-oxo-1,2-dihydropyridine-3-carbonyl)amino]-N-(oxetan-3-yl)-2-phenyl-1H-benzimidazole-6-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Bleicher, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6DB0
 
 | | N-Terminal bromodomain of Human BRD2 with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S,4R)-4-[(2-chlorophenyl)amino]-2-methyl-6-(1H-pyrazol-3-yl)-3,4-dihydroquinolin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | N-Terminal bromodomain of Human BRD2 with a Tetrahydroquinoline analogue
To Be Published
|
|
5SF7
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c2c(ccc1OC)C(=O)N(c3c(nc(n23)c4c(Cl)cccc4)C)C, micromolar IC50=0.0075033 | | Descriptor: | (10S)-1-(2-chlorophenyl)-8-methoxy-3,4-dimethylimidazo[1,5-a]quinazolin-5(4H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6T1G
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
5SGM
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c4(c(C(Nc2cc1nc(cn1cc2)c3ccc(cc3)F)=O)n(nc4)C)C(=O)N(C)C, micromolar IC50=0.000250 | | Descriptor: | MAGNESIUM ION, N~5~-[(4R)-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-7-yl]-N~4~,N~4~,1-trimethyl-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4QWP
 
 | | co-crystal structure of chitosanase OU01 with substrate | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lyu, Q, Liu, W, Han, B. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01.
Biochim.Biophys.Acta, 1850, 2015
|
|
5BVJ
 
 | |
4IOR
 
 | |
5J1Y
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 1-(pyrrolidin-1-yl)-3-(tetrahydrofuran-3-yl)propan-1-one at 1.81A resolution | | Descriptor: | 3-[(3S)-oxolan-3-yl]-1-(pyrrolidin-1-yl)propan-1-one, EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
