8HG1
 
 | | The structure of MPXV polymerase holoenzyme in replicating state | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Peng, Q, Xie, Y.F, Kuai, L, Wang, H, Qi, J.X, Gao, F, Shi, Y. | | Deposit date: | 2022-11-13 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of monkeypox virus DNA polymerase holoenzyme.
Science, 379, 2023
|
|
3TOE
 
 | | Structure of Mth10b | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Pan, X.M, Zhang, N, Liu, Y.F, Liu, X. | | Deposit date: | 2011-09-05 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA.
Plos One, 7, 2012
|
|
7JY9
 
 | | Structure of a 9 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (42-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
8DW1
 
 | | Crystal structure of a host-guest complex with 5'-CTTAGTTATAACTAAG-3' | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*CP*TP*AP*AP*G)-3'), reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-07-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Bioorg.Med.Chem., 77, 2023
|
|
7JY7
 
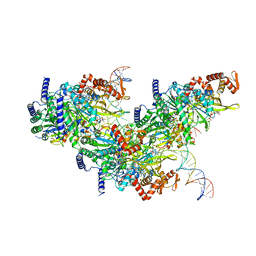 | | Structure of a 12 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (48-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
5JRB
 
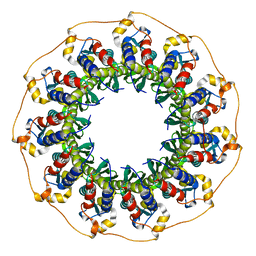 | | Rad52(1-212) K102A/K133A/E202A mutant | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Saotome, M, Saito, K, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2016-05-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the human DNA-repair protein RAD52 containing surface mutations.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3EI4
 
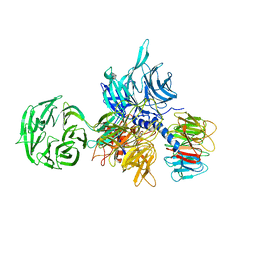 | | Structure of the hsDDB1-hsDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Scrima, A, Pavletich, N.P, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
4QJN
 
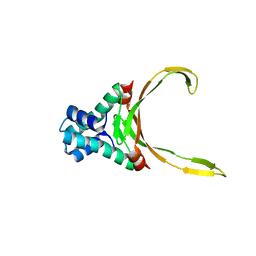 | | Crystal structure of apo nucleoid associated protein, SAV1473 | | Descriptor: | DNA-binding protein HU | | Authors: | Lee, B.-J, Kim, D.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | beta-Arm flexibility of HU from Staphylococcus aureus dictates the DNA-binding and recognition mechanism
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5JZC
 
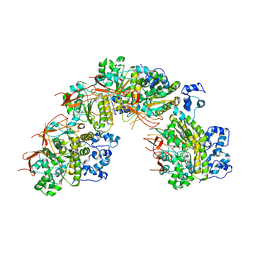 | | helical filament | | Descriptor: | DNA repair protein RAD51 homolog 1 | | Authors: | Short, J, Liu, Y. | | Deposit date: | 2016-05-16 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy.
Nucleic Acids Res., 44, 2016
|
|
1HUU
 
 | |
8CW2
 
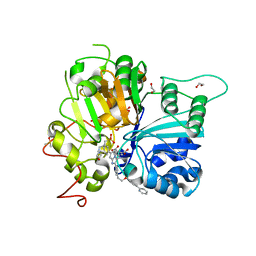 | | Crystal structure of TDP1 complexed with compound XZ760 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-({(4R)-7-phenyl-2-[4-(2-{[4-(pyridin-2-yl)phenyl]methoxy}ethyl)phenyl]imidazo[1,2-a]pyridin-3-yl}amino)benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification.
Rsc Chem Biol, 4, 2023
|
|
8CVQ
 
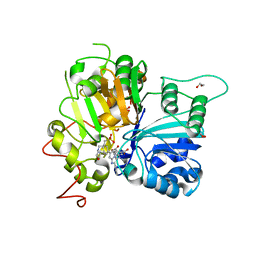 | | Crystal structure of TDP1 complexed with compound XZ761 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification.
Rsc Chem Biol, 4, 2023
|
|
5AH2
 
 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | DNA POLYMERASE III SUBUNIT BETA, GRISELIMYCIN, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
7OGS
 
 | |
8HM0
 
 | | F8-A22-E4 complex of MPXV in trimeric form | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Li, Y.N, Shen, Y.P, Hu, Z.W, Yan, R.H. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the assembly of the DNA polymerase holoenzyme from a monkeypox virus variant.
Sci Adv, 9, 2023
|
|
8HLZ
 
 | | F8-A22-E4 complex of MPXV in hexameric form | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Li, Y.N, Shen, Y.P, Hu, Z.W, Yan, R.H. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the assembly of the DNA polymerase holoenzyme from a monkeypox virus variant.
Sci Adv, 9, 2023
|
|
5JXY
 
 | | Enzyme-substrate complex of TDG catalytic domain bound to a G/U analog | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pidugu, L.S, Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
7OOT
 
 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element | | Descriptor: | DNA (5'-D(P*AP*GP*CP*TP*TP*TP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*GP*TP*TP*G)-3'), DNA (5'-D(P*TP*CP*AP*AP*CP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor 4, ... | | Authors: | Agnarelli, A, El Omari, K, Alt, A.O, Mancini, E.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to interferon-stimulated response element
To Be Published
|
|
3SAV
 
 | | MUTM Slanted complex 8 | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*AP*AP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*T*CP*CP*TP*TP*GP*TP*(CX2)P*TP*AP*CP*C)-3'), DNA GLYCOSYLASE, ... | | Authors: | Spong, M.C, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.125 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FX4
 
 | |
7WB3
 
 | | Crystal structure of T. maritima Rex in ternary complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*GP*AP*GP*AP*AP*AP*TP*TP*TP*AP*TP*CP*AP*CP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*TP*GP*AP*TP*AP*AP*AP*TP*TP*TP*CP*TP*CP*AP*AP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lee, J.Y, Jeong, K.H, Lee, H.J, Park, Y.W. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis of Redox-Sensing Transcriptional Repressor Rex with Cofactor NAD + and Operator DNA.
Int J Mol Sci, 23, 2022
|
|
8DK2
 
 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetABC in an unclamped state trapped in ATP dependent dimeric form | | Descriptor: | DNA (26-MER), JetA, JetB, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
7UB2
 
 | |
7UFZ
 
 | | Crystal structure of TDP1 complexed with compound XZ768 | | Descriptor: | (4-{[(4S)-2-phenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)phosphonic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
