3EUX
 
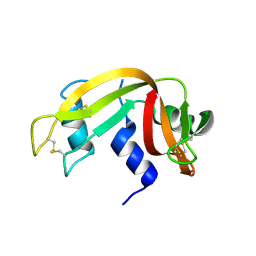 | | Crystal Structure of Crosslinked Ribonuclease A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV4
 
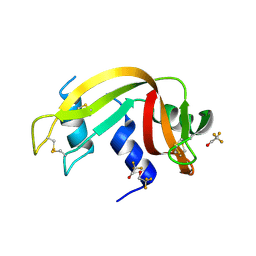 | | Crystal Structure of Ribonuclease A in 50% Trifluoroethanol | | Descriptor: | Ribonuclease pancreatic, TRIFLUOROETHANOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
5Z0A
 
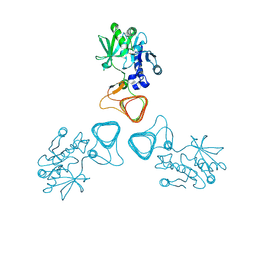 | | ST0452(Y97N)-GlcNAc binding form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual sugar-1-phosphate nucleotidylyltransferase | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
3TPL
 
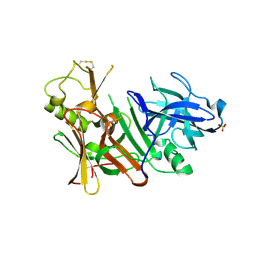 | | APO Structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPR
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3NRC
 
 | | Crystal Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD+ and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (NADH), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD(+) and triclosan.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5F3H
 
 | | Structure of myostatin in complex with humanized RK35 antibody | | Descriptor: | Growth/differentiation factor 8, humanized RK35 antibody heavy chain, humanized RK35 antibody light chain | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Mabs, 8, 2016
|
|
5F3B
 
 | | Structure of myostatin in complex with chimeric RK35 antibody | | Descriptor: | GLYCEROL, Growth/differentiation factor 8, RK35 Chimeric antibody heavy chain, ... | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Mabs, 8, 2016
|
|
6T62
 
 | |
5Z37
 
 | |
6DSS
 
 | | Re-refinement of P. falciparum orotidine 5'-monophosphate decarboxylase | | Descriptor: | Orotidine 5'-monophosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Brandt, G.S, Novak, W.R.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Re-refinement of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase provides a clearer picture of an important malarial drug target.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6DSQ
 
 | |
8WQ9
 
 | |
5ZS9
 
 | |
5EUJ
 
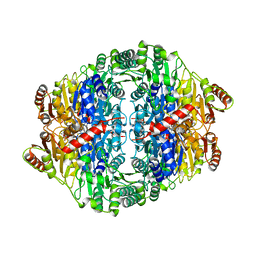 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6DSR
 
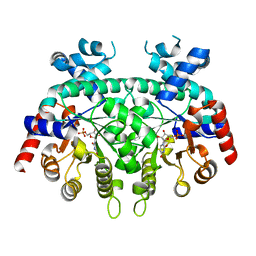 | | Re-refinement of P. falciparum orotidine 5'-monophosphate decarboxylase | | Descriptor: | Orotidine 5'-monophosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Brandt, G.S, Novak, W.R.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Re-refinement of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase provides a clearer picture of an important malarial drug target.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
9G8K
 
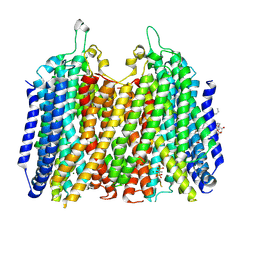 | | Structure of K+-dependent Na+-PPase from Thermotoga maritima in complex with Ca2+ and Etidronate | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Vidilaseris, K, Liu, J, Goldman, A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformational dynamics and asymmetry in multimodal inhibition of membrane-bound pyrophosphatases
Elife, 2024
|
|
8WZU
 
 | |
6T77
 
 | | Crystal structure of Klebsiella pneumoniae FabG(NADPH-dependent) NADP-complex at 1.75 A resolution | | Descriptor: | 3-oxoacyl-ACP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
8UEL
 
 | | Crystal structure of enolase from Litopenaeus vannamei | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Chang, X, Zhao, G. | | Deposit date: | 2023-10-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and Structural Analyses of Enolase from Shrimp ( Litopenaeus vannamei ).
J.Agric.Food Chem., 71, 2023
|
|
6DXQ
 
 | | Crystal structure of the LigJ Hydratase product complex with 4-carboxy-4-hydroxy-2-oxoadipate | | Descriptor: | (2S)-2-hydroxy-4-oxobutane-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
5ZST
 
 | |
7U9K
 
 | | Staphylococcus aureus D-alanine-D-alanine ligase in complex with ATP, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design and synthesis of ATP-competitive N-acyl-substituted sulfamide d-alanine-d-alanine ligase inhibitors.
Bioorg.Med.Chem., 96, 2023
|
|
5ZSK
 
 | |
5ZSP
 
 | | NifS from Hydrogenimonas thermophila | | Descriptor: | Cysteine desulfurase | | Authors: | Nakamura, R, Fujishiro, T, Takahashi, Y. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
