6QRA
 
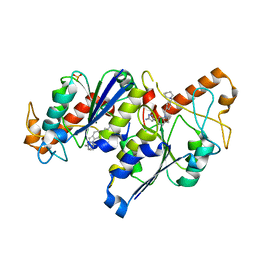 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-[1-[[4-(piperidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazol-5-amine, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
1ZKD
 
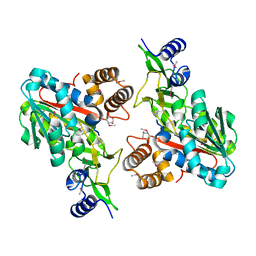 | | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58 | | Descriptor: | DUF185 | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58
To be Published
|
|
6LUD
 
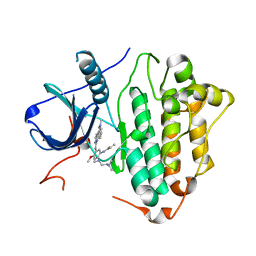 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with Osimertinib | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | Deposit date: | 2020-01-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
5V5G
 
 | |
8JD5
 
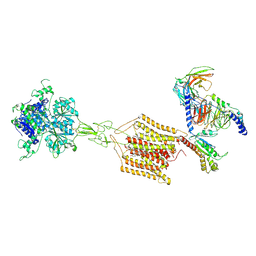 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu4 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-N-(4-methylpyrimidin-2-yl)-4-(1H-pyrazol-4-yl)-1,3-thiazol-2-amine, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8ENT
 
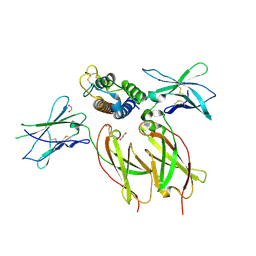 | | Interleukin-21 signaling complex with IL-21R and IL-2Rg | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abhiraman, G.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A structural blueprint for interleukin-21 signal modulation.
Cell Rep, 42, 2023
|
|
8BUS
 
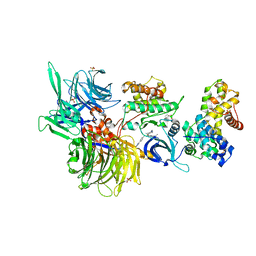 | | Structure of DDB1 bound to DS59-engaged CDK12-cyclin K | | Descriptor: | 1,3-dimethyl-5-[[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)methylamino]purin-2-yl]amino]methyl]pyrazole-4-sulfonamide, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
9HH2
 
 | | Crystal structure of the family S1_19 carrageenan sulfatase ZgCgsA from Zobellia galactanivorans in complex with hybrid a-i-neocarratetraose | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Chevenier, A, Czjzek, M, Michel, G, Ficko-Blean, E. | | Deposit date: | 2024-11-20 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure, function and catalytic mechanism of the carrageenan-sulfatases from the marine bacterium Zobellia galactanivorans Dsij T.
Carbohydr Polym, 358, 2025
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
7H4S
 
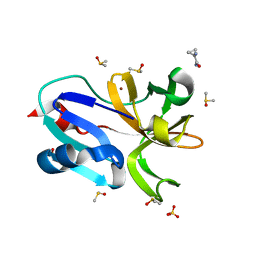 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1415893881 (A71EV2A-x1084) | | Descriptor: | 1-aminocyclopropane-1-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
2R3C
 
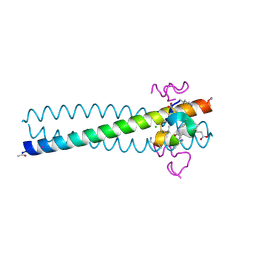 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | Descriptor: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
9HGM
 
 | | Structure of human DNMT2 with bound allosteric inhibitor | | Descriptor: | 4-ethyl-~{N}-[[1-[[(2~{R})-1-(1~{H}-indol-3-yl)-4-[[2-(methylamino)-2-oxidanylidene-ethyl]amino]-4-oxidanylidene-butan-2-yl]carbamoyl]cyclopropyl]methyl]benzamide, GLYCEROL, tRNA (cytosine(38)-C(5))-methyltransferase | | Authors: | Schwan, M, Kopp, J, Sinning, I. | | Deposit date: | 2024-11-20 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA-encoded library screening uncovers potent DNMT2 inhibitors targeting a cryptic allosteric binding site.
Iscience, 28, 2025
|
|
6QIK
 
 | |
4GG7
 
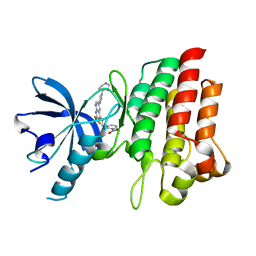 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5WCF
 
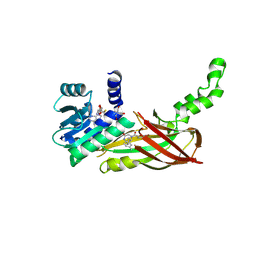 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | (5R)-4-(5-bromofuran-2-carbonyl)-5-(4-fluorophenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Zeng, H, Hutch, A, Seitova, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with MTLLE1441
to be published
|
|
3HVH
 
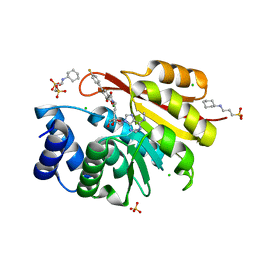 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-methyladenine-containing bisubstrate inhibitor | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7BVU
 
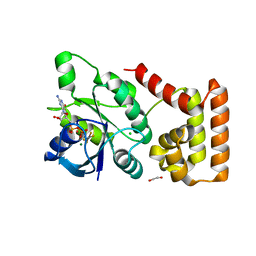 | |
5K44
 
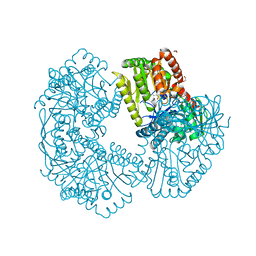 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a complex with Trehalose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
3I2B
 
 | | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase | | Descriptor: | 1,2-ETHANEDIOL, 6-pyruvoyl tetrahydrobiopterin synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ugochukwu, E, Cocking, R, Pilka, E, Yue, W.W, Bray, J.E, Chaikuad, A, Krojer, T, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase
To be Published
|
|
7H81
 
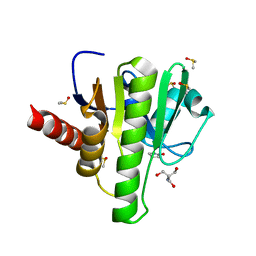 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z940713508 (CHIKV_MacB-x1114) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
4CTS
 
 | |
3DYG
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
6DQV
 
 | | Class 2 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6QEX
 
 | |
6QFE
 
 | | Crystal Structure of Human Kallikrein 5 in complex with GSK144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
