7B1Y
 
 | |
7AS0
 
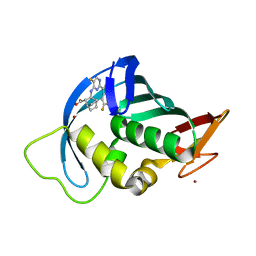 | | Influenza A PB2 in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, BROMIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS1
 
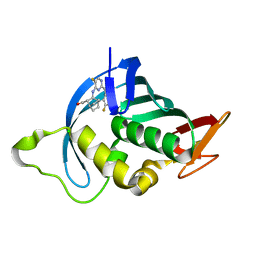 | | Influenza A PB2 (F404Y mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, CHLORIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS2
 
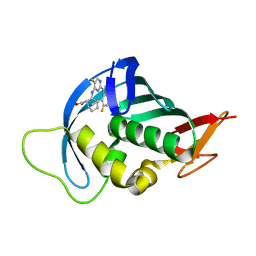 | | Influenza A PB2 (M431 mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS3
 
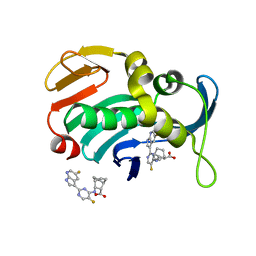 | | Influenza A PB2 (H357N mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7B7R
 
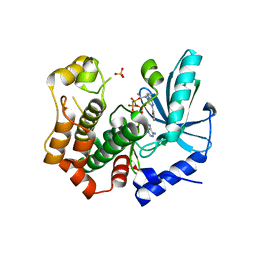 | | MEK1 in complex with compound 4 | | Descriptor: | 2-[5-[ethyl(methyl)amino]imidazo[1,2-a]pyrimidin-7-yl]phenol, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7AVX
 
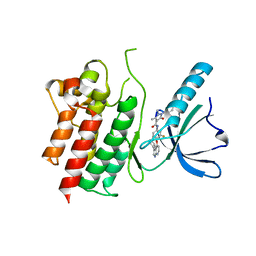 | | MerTK kinase domain in complex with NPS-1034 | | Descriptor: | 1-(4-fluorophenyl)-N-[3-fluoro-4-[(3-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)oxy]phenyl]-2,3-dimethyl-5-oxopyrazole-4-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7B23
 
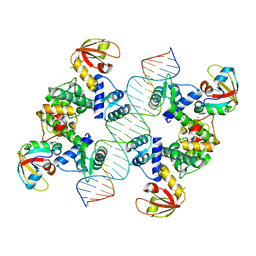 | |
7B3M
 
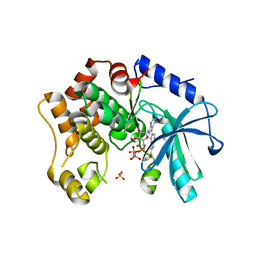 | | MEK1 in complex with compound 6 | | Descriptor: | 1~{H}-indol-2-yl(pyridin-3-yl)methanone, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7AW0
 
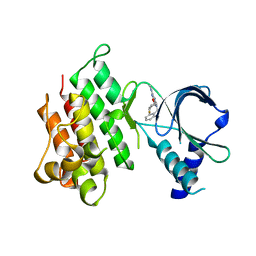 | | MerTK kinase domain in complex with purine inhibitor | | Descriptor: | 2-(cyclopentyloxy)-9-(2,6-difluorobenzyl)-N-methyl-9H-purin-6-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Clarke, M, Disch, J, Goldberg, K, Guilinger, J, Hennessy, E.J, Jetson, R, Ginkunja, D, Hardaker, E, Keefe, A, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S, Zhang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7B1V
 
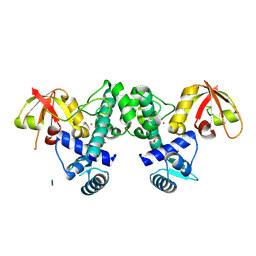 | |
7ALR
 
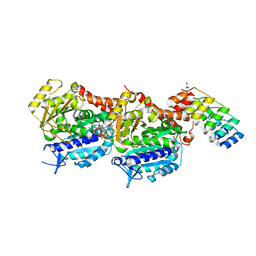 | | Crystal structure of TD1-gatorbulin1 complex | | Descriptor: | (2~{R})-2-oxidanyl-2-[(6~{S},9~{S},12~{S},15~{S},17~{S})-6,10,12,17-tetramethyl-3-methylidene-7-oxidanyl-2,5,8,11,14-pentakis(oxidanylidene)-13-oxa-1,4,7,10-tetrazabicyclo[13.3.0]octadecan-9-yl]ethanamide, Designed Ankyrin Repeat Protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Oliva, M.A, Diaz, J.F. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7B25
 
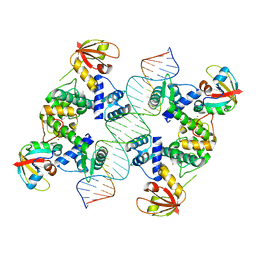 | |
7AW4
 
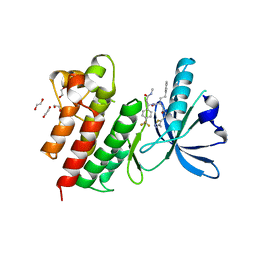 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
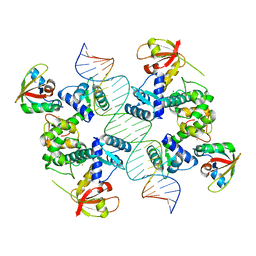
7B20
 
 | |
7AW3
 
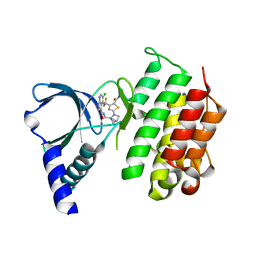 | | MerTK kinase domain with type 1 inhibitor from a DNA-encoded library | | Descriptor: | 2-(1-((5-chloro-1H-pyrrolo[2,3-b]pyridine-3-carboxamido)methyl)-2-azabicyclo[2.1.1]hexan-2-yl)-N-methyl-4-(trifluoromethyl)thiazole-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AHP
 
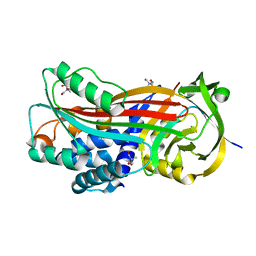 | | Crystal structure of Ixodes ricinus serpin - Iripin-3 | | Descriptor: | Putative salivary serpin, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Kascakova, B, Kuta Smatanova, I, Prudnikova, T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Iripin-3, a New Salivary Protein Isolated From Ixodes ricinus Ticks, Displays Immunomodulatory and Anti-Hemostatic Properties In Vitro
Front Immunol, 12, 2021
|
|
8A5U
 
 | |
6PX5
 
 | | CRYSTAL STRUCTURE OF HUMAN MEIZOTHROMBIN DESF1 MUTANT S195A bound with PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Gistover, N, Di Cera, E. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
2BW2
 
 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
2BVQ
 
 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, HIV-P24, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
6ZRP
 
 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, De Luca, F, Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S, Doquier, J.D. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
2BVU
 
 | |
2BEY
 
 | |
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
