7OZH
 
 | |
7OZG
 
 | |
8AZ6
 
 | | IAPP S20G plateau-phase fibril polymorph 4PF-LU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ1
 
 | | IAPP S20G growth-phase fibril polymorph 2PF-C | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ2
 
 | | IAPP S20G growth-phase fibril polymorph 3PF-CU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
7QBN
 
 | | Structure of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBM
 
 | | Structure of the activation intermediate of cathepsin K in complex with the 3-cyano-3-aza-beta-amino acid inhibitor Gu2602 | | Descriptor: | ACETATE ION, Cathepsin K, MAGNESIUM ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBL
 
 | |
7QBO
 
 | | Structure of the activation intermediate of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8BU0
 
 | | Crystal structure of an 8 repeat consensus TPR superhelix with calcium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CHY
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Zinc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CH0
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Gadolinium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CIG
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix in tris Buffer with Calcium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
7AL0
 
 | | Crystal Structure of Heymonin, a Novel Frog-derived Peptide | | Descriptor: | CHLORIDE ION, Heymonin | | Authors: | Kascakova, B, Prudnikova, T, Kuta Smatanova, I, Xu, X. | | Deposit date: | 2020-10-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization and functional analysis of cathelicidin-MH, a novel frog-derived peptide with anti-septicemic properties.
Elife, 10, 2021
|
|
8OT7
 
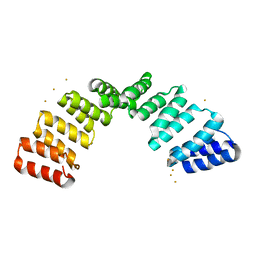 | |
5BXN
 
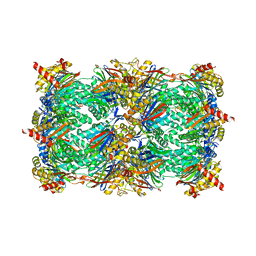 | | Yeast 20S proteasome beta2-G170A mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
5BXL
 
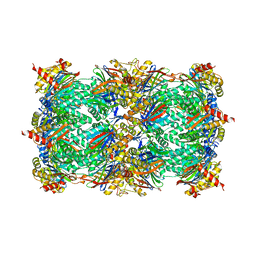 | | Yeast 20S proteasome beta2-G170A mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
4W5P
 
 | | Prp peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4W71
 
 | | Crystal structure of a prion peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-21 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4W5M
 
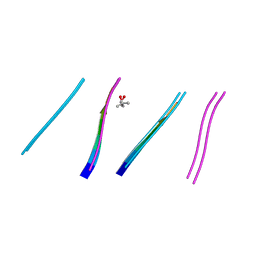 | | Prp peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4W5Y
 
 | | Crystal structure of Prp pepttide | | Descriptor: | Prp peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-19 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.122 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
8AZ0
 
 | | IAPP S20G growth-phase fibril polymorph 2PF-L | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ4
 
 | | IAPP S20G plateau-phase fibril polymorph 2PF-L | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
6V5T
 
 | |
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
