2H5M
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus complexed with acetyl-CoA. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family | | Authors: | Cort, J.R, Ramelot, T.A, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins
J.Struct.Funct.Genom., 9, 2008
|
|
2NOV
 
 | | Breakage-reunion domain of S.pneumoniae topo IV: crystal structure of a gram-positive quinolone target | | Descriptor: | DNA topoisomerase 4 subunit A | | Authors: | Laponogov, I, Veselkov, D.A, Sohi, M.K, Pan, X.S, Achari, A, Yang, C, Ferrara, J.D, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target.
PLoS ONE, 2, 2007
|
|
2NEO
 
 | | SOLUTION NMR STRUCTURE OF A TWO-BASE DNA BULGE COMPLEXED WITH AN ENEDIYNE CLEAVING ANALOG, 11 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*AP*TP*GP*CP*PGE*GP*CP*AP*AP*TP*TP*CP*GP*GP*G)-3'), SPIRO[[7-METHOXY-5-METHYL-1,2-DIHYDRO-NAPHTHALENE]-3,1'-[5-HYDROXY-9-[2-METHYLAMINO-2,6-DIDEOXYGALACTOPYRANOSYL-OXY]-5-(2-OXO-[1,3]DIOXOLAN-4-YL)-3A,5,9,9A-TETRAHYDRO-3H-1-OXA-CYCLOPENTA[A]-S-INDACEN-2-ONE]] | | Authors: | Stassinopoulos, A, Ji, J, Gao, X, Goldberg, I.H. | | Deposit date: | 1997-06-19 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-base DNA bulge complexed with an enediyne cleaving analog.
Science, 272, 1996
|
|
1RTA
 
 | |
1OIQ
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)-2-PYRIMIDINYL]ACETAMIDE | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1OIR
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 1-(DIMETHYLAMINO)-3-(4-{{4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)PYRIMIDIN-2-YL]AMINO}PHENOXY)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
2FXU
 
 | | X-ray Structure of Bistramide A- Actin Complex at 1.35 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rizvi, S.A, Tereshko, V, Kossiakoff, A.A, Kozmin, S.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of bistramide a-actin complex at a 1.35 A resolution
J.Am.Chem.Soc., 128, 2006
|
|
2O01
 
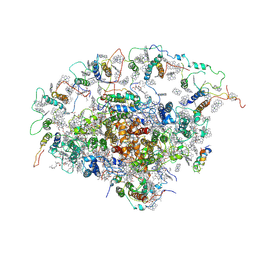 | | The Structure of a plant photosystem I supercomplex at 3.4 Angstrom resolution | | Descriptor: | AT3g54890, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Amunts, A, Drory, O, Nelson, N. | | Deposit date: | 2006-11-27 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structure of a plant photosystem I supercomplex at 3.4 A resolution.
Nature, 447, 2007
|
|
4UWN
 
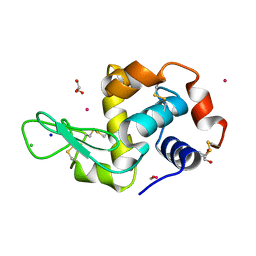 | | Lysozyme soaked with a ruthenium based CORM with a methione oxide ligand (complex 6b) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
3CWO
 
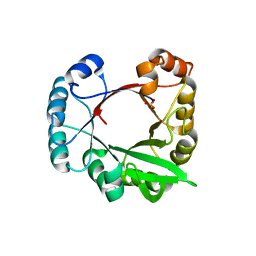 | | A beta/alpha-barrel built by the combination of fragments from different folds | | Descriptor: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | Authors: | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CIQ
 
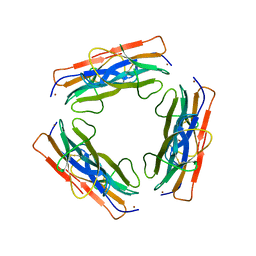 | | A regulatable switch mediates self-association in an immunoglobulin fold | | Descriptor: | Beta-2-microglobulin, COPPER (II) ION, L(+)-TARTARIC ACID | | Authors: | Calabrese, M.F, Eakin, C.M, Wang, J.M, Miranker, A.D. | | Deposit date: | 2008-03-11 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A regulatable switch mediates self-association in an immunoglobulin fold.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1L84
 
 | |
4UWU
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 7) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
2UZS
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
3S0M
 
 | | A Structural Element that Modulates Proton-Coupled Electron Transfer in Oxalate Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saylor, B.T, Reinhardt, L.A, Lu, Z, Shukla, M.S, Cleland, W.W, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2011-05-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase.
Biochemistry, 51, 2012
|
|
1LQT
 
 | | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
6F90
 
 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1U3N
 
 | | A SOD-like protein from B. subtilis, unstructured in solution, becomes ordered in the crystal: implications for function and for fibrillogenesis | | Descriptor: | Hypothetical superoxide dismutase-like protein yojM | | Authors: | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | Deposit date: | 2004-07-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6Y3H
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0401 | | Descriptor: | Major allergen Cor a 1.0401 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3L
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0404 | | Descriptor: | Major allergen variant Cor a 1.0404 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3I
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0402 | | Descriptor: | Major allergen variant Cor a 1.0402 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3K
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | Descriptor: | Major allergen variant Cor a 1.0403 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
6E7H
 
 | |
