6BQC
 
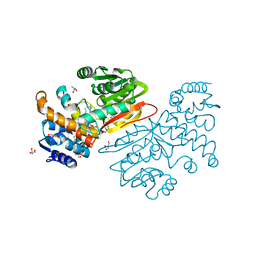 | | Cyclopropane fatty acid synthase from E. coli | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CARBONATE ION, Cyclopropane-fatty-acyl-phospholipid synthase, ... | | Authors: | Hari, S.B, Grant, R.A, Sauer, R.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural and Functional Analysis of E. coli Cyclopropane Fatty Acid Synthase.
Structure, 26, 2018
|
|
6R2L
 
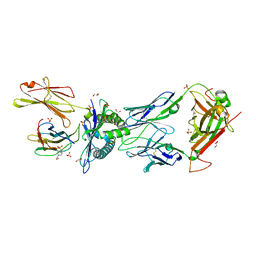 | | NYBR1-A2-SLSKILDTV | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sami, M. | | Deposit date: | 2019-03-18 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity of bispecific T cell receptors and antibodies targeting peptide-HLA.
J.Clin.Invest., 130, 2020
|
|
8ENI
 
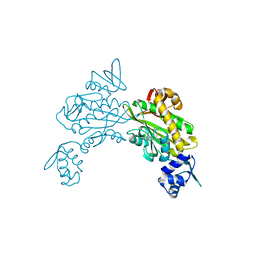 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Wilce, M.C.J, Cini, D.A. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6BQL
 
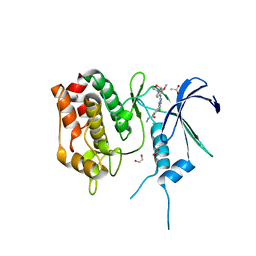 | | Crystal Structure of the Human CAMKK2B in complex with TAE-226 | | Descriptor: | 1,2-ETHANEDIOL, 2-({5-CHLORO-2-[(2-METHOXY-4-MORPHOLIN-4-YLPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)-N-METHYLBENZAMIDE, ACETATE ION, ... | | Authors: | Counago, R.M, de Souza, G.P, dos Reis, C.V, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with TAE-226
To Be Published
|
|
4KRT
 
 | |
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | Descriptor: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
4ZY0
 
 | |
5VW7
 
 | | NADPH soak of Y316A mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
6BVT
 
 | | Importin alpha 1 in cargo free state | | Descriptor: | Importin subunit alpha-1 | | Authors: | Smith, K.M, Tsimablyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6U37
 
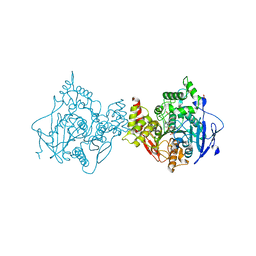 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6F8R
 
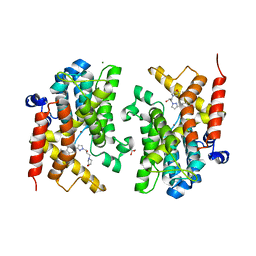 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-54 | | Descriptor: | (2~{S})-1-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
5DMR
 
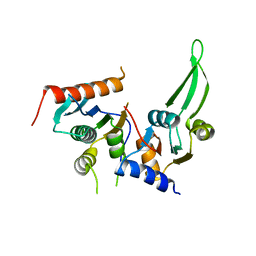 | |
4CDW
 
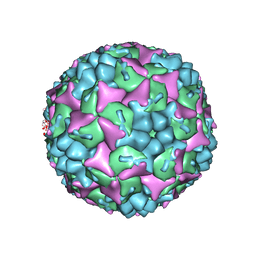 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
8ZH3
 
 | | 80S ribosome with A/P-P/E tRNA and mRNA of WNV | | Descriptor: | 25S rRNA (3393-MER), 5.8S rRNA (158-MER), 5S rRNA (121-MER), ... | | Authors: | Wu, M, Yuan, S. | | Deposit date: | 2024-05-10 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Flexibilities and conservations in the structural basis of viral -1 programmed ribosomal frameshifting
To Be Published
|
|
8ZHB
 
 | | 80S ribosome with A/A P/E tRNA and mRNA of WNV | | Descriptor: | 25S rRNA (3393-MER), 5.8S rRNA (158-MER), 5S rRNA (121-MER), ... | | Authors: | Wu, M, Yuan, S. | | Deposit date: | 2024-05-10 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexibilities and conservations in the structural basis of viral -1 programmed ribosomal frameshifting
To Be Published
|
|
7KKE
 
 | | Phosphoinositide 3-Kinase gamma bound to a thiazole inhibitor | | Descriptor: | N-[2-(3,3-dimethylbutoxy)ethyl]-N'-{4-methyl-5-[(pyridin-4-yl)ethynyl]-1,3-thiazol-2-yl}urea, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Jacobs, M.D, Griffith, J.P. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Novel Series of Potent and Selective Alkynylthiazole-Derived PI3K gamma Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6U4O
 
 | |
1YSL
 
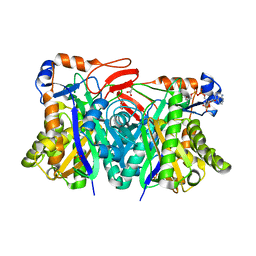 | | Crystal structure of HMG-CoA synthase from Enterococcus faecalis with AcetoAcetyl-CoA ligand. | | Descriptor: | ACETOACETIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Steussy, C.N, Vartia, A.A, Burgner II, J.W, Sutherlin, A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2005-02-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structures of HMG-CoA Synthase from Enterococcus faecalis and a Complex with Its Second Substrate/Inhibitor Acetoacetyl-CoA.
Biochemistry, 44, 2005
|
|
5W6E
 
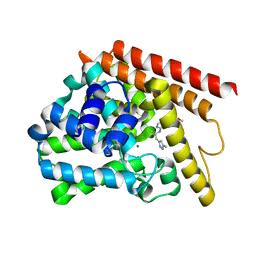 | | PDE1b complexed with compound 3S | | Descriptor: | 7,8-dimethoxy-N-[(2S)-1-(3-methyl-1H-pyrazol-5-yl)propan-2-yl]quinazolin-4-amine, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2017-06-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Periphery-Restricted Quinazoline Inhibitors of the Cyclic Nucleotide Phosphodiesterase PDE1.
J. Med. Chem., 61, 2018
|
|
5TQJ
 
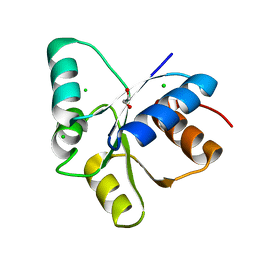 | |
4KJT
 
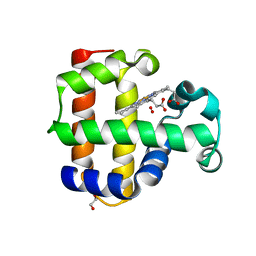 | | Structure of the L100F MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH OXYGEN | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations.
Arch.Biochem.Biophys., 545, 2014
|
|
7KL5
 
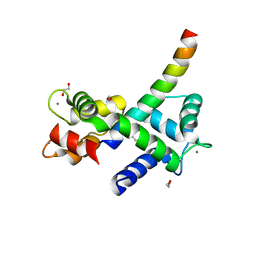 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
7KQ4
 
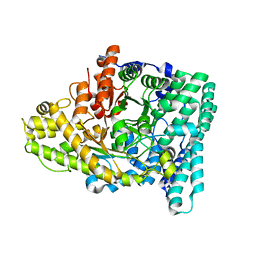 | |
7KJZ
 
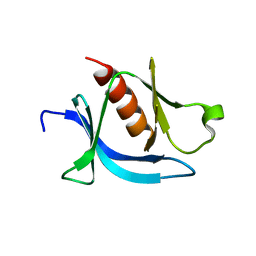 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
8ZGR
 
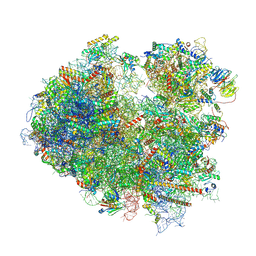 | | 80S ribosome with A/A tRNA and mRNA of WNV | | Descriptor: | 25S rRNA (3393-MER), 5.8S rRNA (158-MER), 5S rRNA (121-MER), ... | | Authors: | Wu, M, Yuan, S. | | Deposit date: | 2024-05-09 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Flexibilities and conservations in the structural basis of viral -1 programmed ribosomal frameshifting
To Be Published
|
|
