8DUK
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-(methylamino)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-{4-[2-(methylamino)ethoxy]phenyl}-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
5K9G
 
 | | Crystal Structure of GTP Cyclohydrolase-IB with Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Alvarez, J, Stec, B, Swairjo, M.A. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB.
Biochem.J., 474, 2017
|
|
8BUS
 
 | | Structure of DDB1 bound to DS59-engaged CDK12-cyclin K | | Descriptor: | 1,3-dimethyl-5-[[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)methylamino]purin-2-yl]amino]methyl]pyrazole-4-sulfonamide, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
9EET
 
 | |
9D7T
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Api137 antimicrobial peptide, mRNA, A-site release factor 1, and deacylated P-site and E-site tRNAphe at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Huang, W, Baliga, C, Atkinson, G.C, Vazquez-Laslop, N, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-17 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activity, structure, and diversity of Type II proline-rich antimicrobial peptides from insects.
Embo Rep., 25, 2024
|
|
9D7R
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Fva1 antimicrobial peptide, mRNA, A-site release factor 1, and deacylated P-site and E-site tRNAphe at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Huang, W, Baliga, C, Atkinson, G.C, Vazquez-Laslop, N, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-17 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activity, structure, and diversity of Type II proline-rich antimicrobial peptides from insects.
Embo Rep., 25, 2024
|
|
5FH1
 
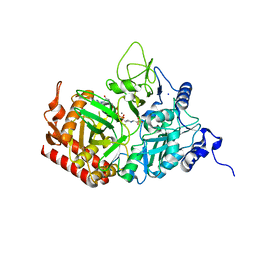 | | The structure of rat cytosolic PEPCK variant E89D in complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
5WNM
 
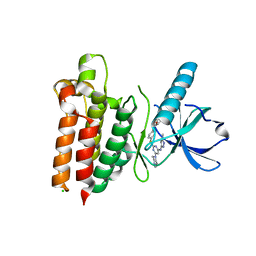 | |
5FPS
 
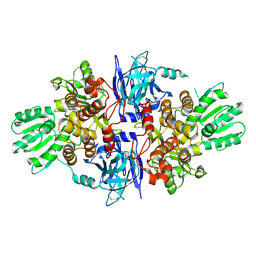 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 3-aminobenzene-1,2-dicarboxylic acid (AT1246) in an alternate binding site. | | Descriptor: | 3-AMINOBENZENE-1,2-DICARBOXYLIC ACID, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CTX
 
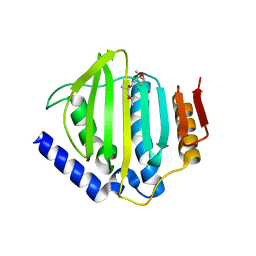 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4RCF
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 4-fluoroxanthene inhibitor 49 | | Descriptor: | (4S)-2'-(3,6-dihydro-2H-pyran-4-yl)-4'-fluoro-7'-(2-fluoropyridin-3-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Lead Optimization and Modulation of hERG Activity in a Series of Aminooxazoline Xanthene beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
5KGJ
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
4QVN
 
 | | yCP beta5-M45V mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
2YBY
 
 | | Structure of domains 6 and 7 of the mouse complement regulator Factor H | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H | | Authors: | Everett, R.J, Caesar, J.J.E, Johnson, S.J, Tang, C.M, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
6HWA
 
 | | Yeast 20S proteasome in complex with 43 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[(4~{a}~{S},8~{a}~{S})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-2-yl]-4-methyl-3,4-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
4REW
 
 | | Crystal structure of the non-phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
6SLO
 
 | | Crystal structure of PUF60 UHM domain in complex with 7,8 dimethoxyperphenazine | | Descriptor: | 2-[4-[3-(8-chloranyl-2,3-dimethoxy-phenothiazin-10-yl)propyl]piperazin-1-yl]ethanol, MAGNESIUM ION, Thioredoxin,Poly(U)-binding-splicing factor PUF60 | | Authors: | Jagtap, P.K.A, Kubelka, T, Bach, T, Sattler, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly.
Nat Commun, 11, 2020
|
|
6SPN
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
8G7R
 
 | |
6QQV
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 2-[[6-(5-azanyl-1~{H}-pyrazol-3-yl)indol-1-yl]methyl]benzenecarbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
6B4W
 
 | | TTK in Complex with Inhibitor | | Descriptor: | 4-{[4-(cyclopentyloxy)-5-(2-methyl-1,3-benzoxazol-6-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}-3-methoxy-N-methylbenzamide, CACODYLATE ION, Dual specificity protein kinase TTK | | Authors: | Delker, S, Chamberlain, P.P. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery of a Dual TTK Protein Kinase/CDC2-Like Kinase (CLK2) Inhibitor for the Treatment of Triple Negative Breast Cancer Initiated from a Phenotypic Screen.
J. Med. Chem., 60, 2017
|
|
5FMJ
 
 | | Bcl-xL with mouse Bak BH3 Q75L complex | | Descriptor: | 1,2-ETHANEDIOL, BAK1 PROTEIN, BCL-2-LIKE PROTEIN 1 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
1AKS
 
 | | CRYSTAL STRUCTURE OF THE FIRST ACTIVE AUTOLYSATE FORM OF THE PORCINE ALPHA TRYPSIN | | Descriptor: | ALPHA TRYPSIN, CALCIUM ION | | Authors: | Johnson, A, Krishnaswamy, S, Sundaram, P.V, Pattabhi, V. | | Deposit date: | 1996-07-24 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure at 1.8 A resolution of an active autolysate form of porcine alpha-trysoin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1Q93
 
 | | Crystal structure of a mutant of the sarcin/ricin domain from rat 28S rRNA | | Descriptor: | SODIUM ION, SULFATE ION, Sarcin/Ricin 28S rRNA | | Authors: | Correll, C.C, Beneken, J, Plantinga, M.J, Lubbers, M, Chan, Y.L. | | Deposit date: | 2003-08-22 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The common and distinctive features of the bulged-G motif based on a 1.04 A resolution RNA structure
Nucleic Acids Res., 31, 2003
|
|
6QR9
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(morpholin-4-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
