6DD0
 
 | | Crystal structure of VIM-2 complexed with compound 8 | | Descriptor: | ACETATE ION, Beta-lactamase class B VIM-2, ZINC ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
5GHV
 
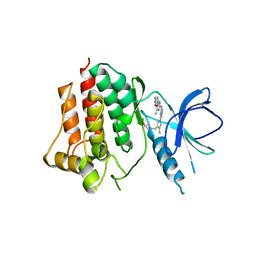 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
6DDJ
 
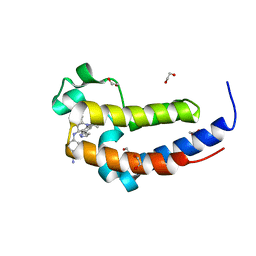 | | Crystal Structure of the human BRD2 BD2 bromodimain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
5ZZC
 
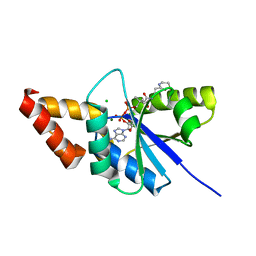 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 1.94A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Gupta, A, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-05-31 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 1.94 A resolution
To Be Published
|
|
4XI0
 
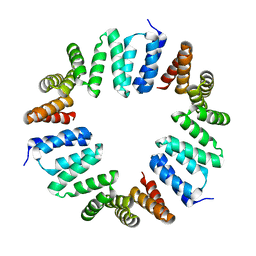 | | MamA 41-end from Desulfovibrio magneticus RS-1 | | Descriptor: | Magnetosome protein MamA | | Authors: | Zarivach, R, Zeytuni, N, Cronin, S, Davidov, G, Baran, D, Stein, T. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MamA as a Model Protein for Structure-Based Insight into the Evolutionary Origins of Magnetotactic Bacteria.
Plos One, 10, 2015
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
4XJ5
 
 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
8R2O
 
 | | Huntingtin-Q17, 1-66, N-MBP fusion | | Descriptor: | Maltodextrin-binding protein,Huntingtin, myristoylated N-terminal fragment, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Toledo-Sherman, L, Dominguez, C. | | Deposit date: | 2023-11-07 | | Release date: | 2024-09-18 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Post-translational modifications of huntingtin's N17 region: implications for self-association and membrane binding.
Biochim Biophys Acta Mol Basis Dis, 1871, 2025
|
|
7X09
 
 | | Homo sapiens Prolyl-tRNA Synthetase (HsPRS) in Complex with inhibitors L95 and Halofuginone | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, BROMIDE ION, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Manickam, Y, Babbar, P, Pillai, P, Sharma, A. | | Deposit date: | 2022-02-21 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Homo sapiens Prolyl-tRNA synthetase (HsPRS) with double inhibitors (HF and L95)
To Be Published
|
|
3GPE
 
 | | Crystal Structure Analysis of PKC (alpha)-C2 domain complexed with Ca2+ and PtdIns(4,5)P2 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Protein kinase C alpha type, ... | | Authors: | Ferrer-Orta, C, Querol-Audi, J, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8R38
 
 | | BIIG2 anti-integrin Fab | | Descriptor: | BIIG2 Fab, heavy chain, light chain, ... | | Authors: | Cordara, G, Heim, J.B, Johannesen, H, Krengel, U. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-25 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Shared ligand-blocking mechanism but distinct conformational modulation by alpha 5-targeting antibodies BIIG2 and MINT1526A.
Biorxiv, 2025
|
|
8AGK
 
 | | Botulinum neurotoxin subtype A6 cell binding domain in complex with GD1a ganglioside | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Newell, A.R, Mojanaga, O.O. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
8DM9
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
5ZU4
 
 | | Crystal structure of Lamprey immune protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Q.W, Pang, Y. | | Deposit date: | 2018-05-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a pore-forming protein
To Be Published
|
|
5NWZ
 
 | |
5I9I
 
 | | Crystal structure of LP_PLA2 in complex with Darapladib | | Descriptor: | N-[2-(diethylamino)ethyl]-2-{2-[(4-fluorobenzyl)sulfanyl]-4-oxo-4,5,6,7-tetrahydro-1H-cyclopenta[d]pyrimidin-1-yl}-N-{[ 4'-(trifluoromethyl)biphenyl-4-yl]methyl}acetamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Characterization of Protein-Ligand Interactions Formed between Lipoprotein-Associated Phospholipase A2 and Inhibitors
J.Med.Chem., 59, 2016
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6SE8
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
3H81
 
 | |
6DNT
 
 | | UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase, ... | | Authors: | Carbone, V, Schofield, L.R, Sang, C, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural determination of archaeal UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with the bacterial cell wall intermediate UDP-N-acetylmuramic acid.
Proteins, 86, 2018
|
|
6DKB
 
 | | Crystal structure of Trk-A in complex with the Pan-Trk Kinase Inhibitor, compound 10b. | | Descriptor: | 2-{[(3R,4S)-3-fluoro-1-{[4-(trifluoromethoxy)phenyl]acetyl}piperidin-4-yl]oxy}-5-(1-methyl-1H-imidazol-4-yl)pyridine-3-carboxamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-05-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Potent, Selective, and Peripherally Restricted Pan-Trk Kinase Inhibitors for the Treatment of Pain.
J. Med. Chem., 61, 2018
|
|
3L7K
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (15 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Strynadka, N.C.J, Lovering, A.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5S36
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2856434938 | | Descriptor: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.058 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6SA2
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX3 (10) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-morpholin-4-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
7KO0
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD4 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Karim, M.R, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
