6F10
 
 | | GLIC mutant D88N | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
6F1T
 
 | | Cryo-EM structure of two dynein tail domains bound to dynactin and BICDR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F4S
 
 | | Human JMJD5 (N308C) in complex with Mn(II), 2OG and RCCD1 (139-143) (complex-4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-OXOGLUTARIC ACID, GLYCEROL, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6EXV
 
 | | Structure of mammalian RNA polymerase II elongation complex inhibited by Alpha-amanitin | | Descriptor: | AMATOXIN, DNA (25-MER), DNA (36-MER), ... | | Authors: | Liu, X, Farnung, L, Wigge, C, Cramer, P. | | Deposit date: | 2017-11-09 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by alpha-amanitin.
J. Biol. Chem., 293, 2018
|
|
6EZN
 
 | | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Kowal, J, Eyring, J, Ngwa, E.M, Aebi, M, Locher, K.P. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
6F0R
 
 | | GLIC mutant E82Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
6F2D
 
 | | A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2017-11-24 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the core of the type III secretion system export apparatus.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F3A
 
 | | Cryo-EM structure of a single dynein tail domain bound to dynactin and BICD2N | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F5V
 
 | | Crystal structure of the prephenate aminotransferase from Arabidopsis thaliana | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Robin, A, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-03 | | Release date: | 2019-03-13 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
5C51
 
 | | Probing the Structural and Molecular Basis of Nucleotide Selectivity by Human Mitochondrial DNA Polymerase gamma | | Descriptor: | DNA, DNA (5'-D(*(AD)P*AP*AP*AP*CP*GP*AP*GP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*GP*TP*AP*C)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Sohl, C.D, Szymanski, M.R, Mislak, A.C, Shumate, C.K, Amiralaei, S, Schinazi, R.F, Anderson, K.S, Whitney, Y.Y. | | Deposit date: | 2015-06-19 | | Release date: | 2016-05-11 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (3.426 Å) | | Cite: | Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C0X
 
 | |
5C3F
 
 | | Crystal structure of Mcl-1 bound to BID-MM | | Descriptor: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
5C5X
 
 | | CRYSTAL STRUCTURE OF THE S156E MUTANT OF HUMAN AQUAPORIN 5 | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Kitchen, P, Oeberg, F, Sjoehamn, J, Hedfalk, K, Bill, R.M, Conner, A.C, Conner, M.T, Toernroth-Horsefield, S. | | Deposit date: | 2015-06-22 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasma Membrane Abundance of Human Aquaporin 5 Is Dynamically Regulated by Multiple Pathways.
Plos One, 10, 2015
|
|
5BW9
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | Authors: | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | Deposit date: | 2015-06-06 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
5C16
 
 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
6ULV
 
 | | BRD4-BD1 in complex with the cyclic peptide 4.2_1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bromodomain-containing protein 4, Cyclic peptide 4.2_3, ... | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-10-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5C52
 
 | | Probing the Structural and Molecular Basis of Nucleotide Selectivity by Human Mitochondrial DNA Polymerase gamma | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*AP*AP*AP*CP*GP*AP*GP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*GP*TP*AP*C)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Sohl, C.D, Szymanski, M.R, Mislak, A.C, Shumate, C.K, Amiralaei, S, Schinazi, R.F, Anderson, K.S, Yin, Y.W. | | Deposit date: | 2015-06-19 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.637 Å) | | Cite: | Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8X
 
 | | ZHD-Intermediate complex after ZHD crystal soaking in ZEN for 20min | | Descriptor: | (1E,10S)-1-(3,5-dihydroxyphenyl)-10-hydroxyundec-1-en-6-one, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Case Study of Lactonase ZHD
To Be Published
|
|
5C42
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K101P) Variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, HIV-1 Reverse Transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Gray, W.T, Anderson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Inhibitors Active against HIV Reverse Transcriptase with K101P, a Mutation Conferring Rilpivirine Resistance.
Acs Med.Chem.Lett., 6, 2015
|
|
5CLW
 
 | | Crystal structure of human glycogen branching enzyme (GBE1) in complex with maltoheptaose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krojer, T, Froese, D.S, Goubin, S, Strain-Damerell, C, Mahajan, P, Burgess-Brown, N, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human glycogen branching enzyme (GBE1) in complex with maltoheptaose
To be published
|
|
5CA2
 
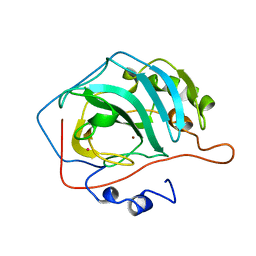 | |
5CAC
 
 | | REFINED STRUCTURE OF HUMAN CARBONIC ANHYDRASE II AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CARBONIC ANHYDRASE FORM C, SULFITE ION, ZINC ION | | Authors: | Lindahl, M, Habash, D, Harrop, S, Helliwell, D.R, Liljas, A. | | Deposit date: | 1991-09-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of human carbonic anhydrase II at 2.0 A resolution.
Proteins, 4, 1988
|
|
5CE3
 
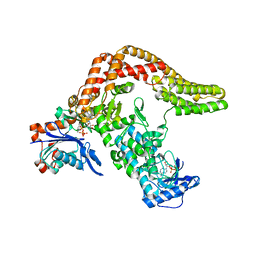 | | The Yersinia YopO - actin complex with MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Lee, W.L, Singaravelu, P, Grimes, J.M, Robinson, R.C. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | The Yersinia YopO - actin complex with MgADP
To Be Published
|
|
8FFZ
 
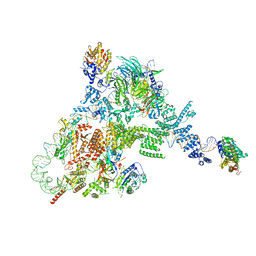 | | TFIIIA-TFIIIC-Brf1-TBP complex bound to 5S rRNA gene | | Descriptor: | DNA (151-MER), Transcription factor IIIA, Transcription factor IIIB 70 kDa subunit,TATA-box-binding protein, ... | | Authors: | Talyzina, A, He, Y. | | Deposit date: | 2022-12-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of TFIIIC-dependent RNA polymerase III transcription initiation.
Mol.Cell, 83, 2023
|
|
6D1W
 
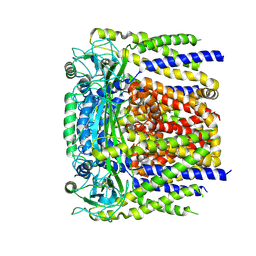 | | human PKD2 F604P mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Zheng, W, Yang, X, Bulkley, D, Chen, X.Z, Cao, E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels.
Nat Commun, 9, 2018
|
|
