6SL6
 
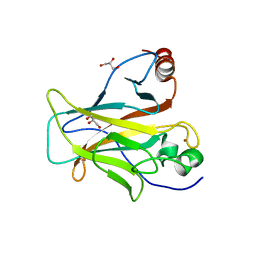 | | p53 charged core | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Gallardo, R, Langenberg, T, Schymkowitz, J, Rousseau, F, Ulens, C. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Thermodynamic and Evolutionary Coupling between the Native and Amyloid State of Globular Proteins.
Cell Rep, 31, 2020
|
|
6SLB
 
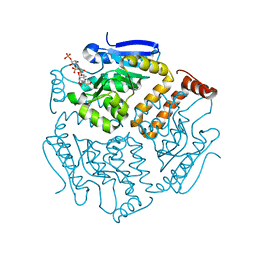 | | Crystal structure of isomerase PaaG with trans-3,4-didehydroadipyl-CoA | | Descriptor: | (~{E})-6-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-6-oxidanylidene-hex-3-enoic acid, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Saleem-Batcha, R, Spieker, M, Teufel, R. | | Deposit date: | 2019-08-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
6MEV
 
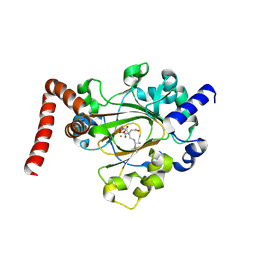 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
4H6E
 
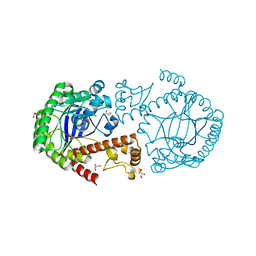 | | tRNA-guanine transglycosylase Y106F, C158V, V233G mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
5GI5
 
 | |
3T27
 
 | | Orthorhombic trypsin (bovine) in the presence of betaine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Venkat, M, Saxby, C, Cahn, J, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6K80
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Complex with CoA and Tryptophol | | Descriptor: | 2-(1H-indol-3-yl)ethanol, ACETYL COENZYME *A, Dopamine N-acetyltransferase | | Authors: | Wu, C.Y, Hu, I.C, Yang, Y.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
5JO0
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC56 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
6AOW
 
 | |
6ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C COMPLEX WITH FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-23 | | Release date: | 2000-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
3T26
 
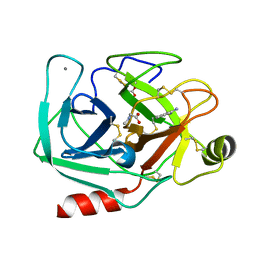 | | Orthorhombic trypsin (bovine) in the presence of sarcosine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Cahn, J, Venkat, M, Marshall, H, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4H7Z
 
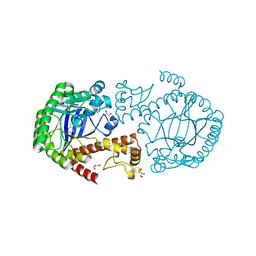 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with guanine | | Descriptor: | GLYCEROL, GUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
6B9F
 
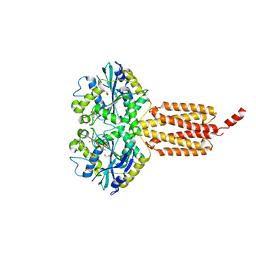 | | Human ATL1 mutant - F151S bound to GDPAlF4- | | Descriptor: | Atlastin-1, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | O'Donnell, J.P, Sondermann, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A hereditary spastic paraplegia-associated atlastin variant exhibits defective allosteric coupling in the catalytic core.
J. Biol. Chem., 293, 2018
|
|
2A6K
 
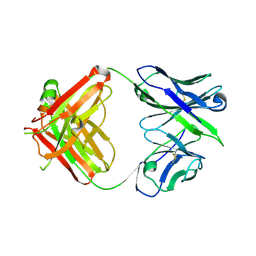 | | Crystal Structure Analysis of the germline antibody 36-65 Fab in complex with the dodecapeptide SLGDNLTNHNLR | | Descriptor: | DODECAPEPTIDE: SLGDNLTNHNLR, Germline antibody 36-65 Fab heavy chain, Germline antibody 36-65 Fab light chain | | Authors: | Sethi, D.K, Agarwal, A, Manivel, V, Rao, K.V, Salunke, D.M. | | Deposit date: | 2005-07-03 | | Release date: | 2006-06-13 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential epitope positioning within the germline antibody paratope enhances promiscuity in the primary immune response.
Immunity, 24, 2006
|
|
4GD0
 
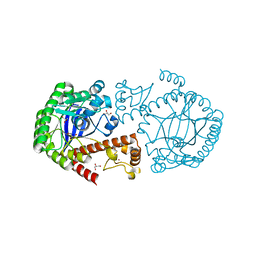 | | tRNA-guanine transglycosylase Y106F, C158V mutant | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Schmidt, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
6MOA
 
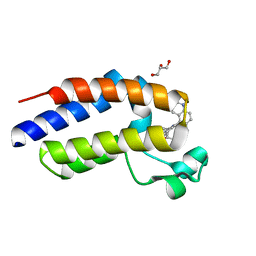 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO7
 
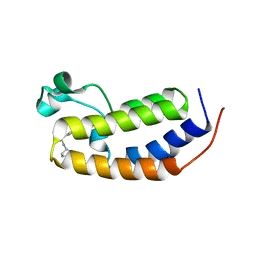 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
3R9A
 
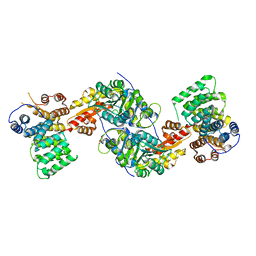 | | Human alanine-glyoxylate aminotransferase in complex with the TPR domain of human PEX5P | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peroxisomal targeting signal 1 receptor, Serine--pyruvate aminotransferase | | Authors: | Fodor, K, Wilmanns, M. | | Deposit date: | 2011-03-25 | | Release date: | 2011-05-11 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1
Plos Biol., 10, 2012
|
|
4HSH
 
 | | tRNA-guanine transglycosylase Y106F, V233G mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4HVX
 
 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-11-07 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
2K4A
 
 | |
3TWL
 
 | | Crystal structure of Arabidopsis thaliana FPG | | Descriptor: | Formamidopyrimidine-DNA glycosylase 1, GLYCEROL | | Authors: | Duclos, S, Aller, P, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-09-22 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine.
Dna Repair, 11, 2012
|
|
3TWK
 
 | | Crystal structure of arabidopsis thaliana FPG | | Descriptor: | Formamidopyrimidine-DNA glycosylase 1, GLYCEROL | | Authors: | Duclos, S, Aller, P, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-09-22 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine.
Dna Repair, 11, 2012
|
|
4HQV
 
 | | tRNA-guanine transglycosylase Y106F, C158V, V233G mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
5FO1
 
 | |
