7KXL
 
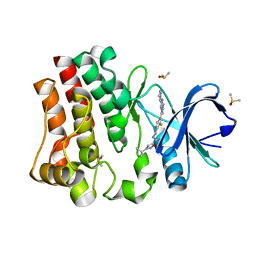 | | BTK1 SOAKED WITH COMPOUND 5, Y551 IS SEQUESTERED | | Descriptor: | 3-tert-butyl-N-({2-fluoro-4-[2-(1-methyl-1H-pyrazol-4-yl)-1H-imidazo[4,5-b]pyridin-7-yl]phenyl}methyl)-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
8FKQ
 
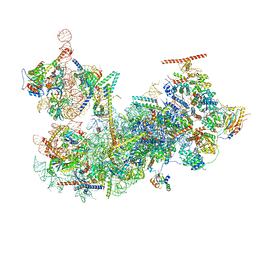 | |
7SUK
 
 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
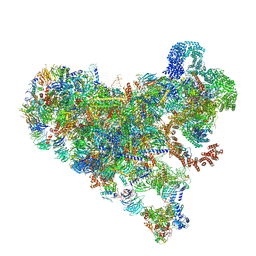
8D80
 
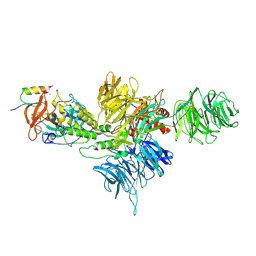 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8SPC
 
 | |
6DWK
 
 | | SAMHD1 Bound to Fludarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-fluoro-9-{5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-a mine, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8JYX
 
 | |
7MOC
 
 | | Neurofibromin core | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9B9G
 
 | | Structure of the PI4KA complex bound to Calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, Hyccin, ... | | Authors: | Shaw, A.L, Suresh, S, Yip, C.K, Burke, J.E. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of calcineurin bound to PI4KA reveals dual interface in both PI4KA and FAM126A.
Structure, 32, 2024
|
|
4TTK
 
 | |
7MP5
 
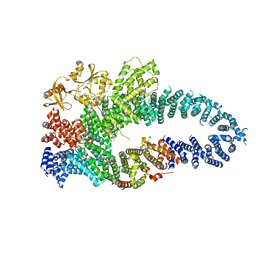 | | Autoinhibited neurofibrobmin | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
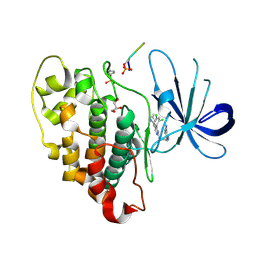
6B8J
 
 | |
6T1O
 
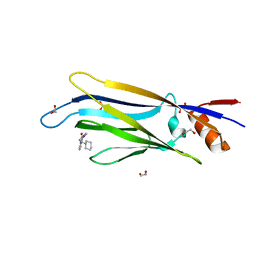 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6P2P
 
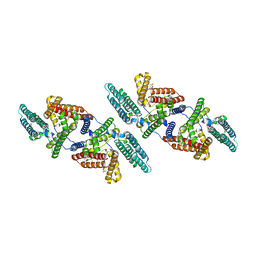 | | Tetrameric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
5HCX
 
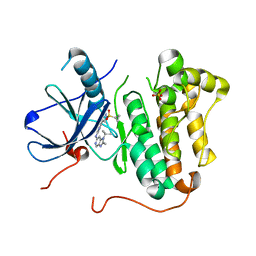 | | EGFR kinase domain mutant "TMLR" with azabenzimidazole compound 7 | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]-2-methyl-1-propan-2-yl-imidazo[4,5-c]pyridin-6-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Noncovalent, Mutant-Selective Epidermal Growth Factor Receptor Inhibitor.
J.Med.Chem., 59, 2016
|
|
7PXY
 
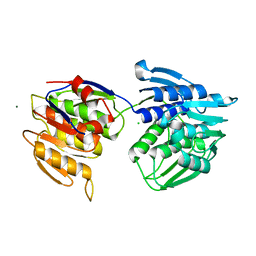 | |
6P3Y
 
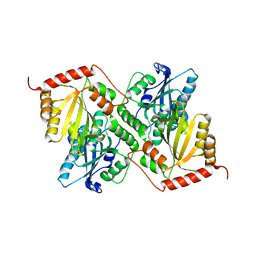 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
5KVW
 
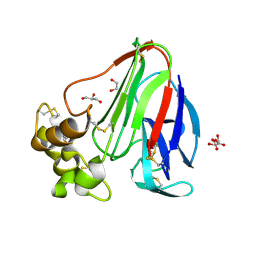 | | T. danielli thaumatin at 100K, Data set 1 | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
4RMH
 
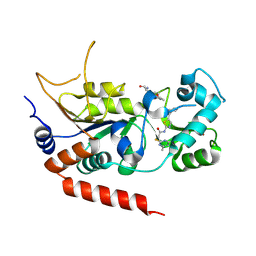 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
9EIL
 
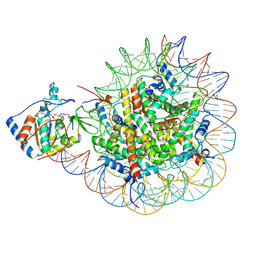 | | SIRT6 bound to an H3K27Ac nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2024-11-26 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and enzymatic plasticity of SIRT6 deacylase activity.
J.Biol.Chem., 301, 2025
|
|
6FKZ
 
 | |
8I34
 
 | | The crystal structure of EPD-BCP1 from a marine sponge | | Descriptor: | (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-4,8,13,17-tetramethyl-3-oxidanyl-19-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]-1-[(1~{R},4~{S})-1,2,2-trimethyl-4-oxidanyl-cyclopentyl]nonadeca-2,4,6,8,10,12,14,16-octaen-18-yn-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASTAXANTHIN, ... | | Authors: | Shomura, Y, Kawasaki, S. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | An ependymin-related blue carotenoprotein decorates marine blue sponge.
J.Biol.Chem., 299, 2023
|
|
9DWV
 
 | | Ternary complex of CRBN-DDB1-PPIL4 RRM domain with FPFT-2216 | | Descriptor: | (3S)-3-[(4M)-4-(4-methoxythiophen-3-yl)-1H-1,2,3-triazol-1-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Peptidyl-prolyl cis-trans isomerase-like 4, ... | | Authors: | Baek, K, Fischer, E.S. | | Deposit date: | 2024-10-10 | | Release date: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unveiling the hidden interactome of CRBN molecular glues.
Nat Commun, 16, 2025
|
|
4RRV
 
 | |
8CRV
 
 | | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Carbamate kinase, FORMIC ACID, ... | | Authors: | Kim, Y, Skarina, T, Mesa, N, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa
To Be Published
|
|
