8EQF
 
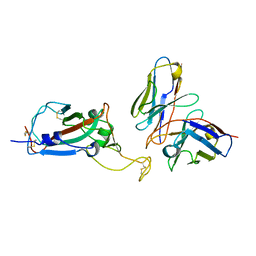 | | cryoEM structure of a broadly neutralizing anti-SARS-CoV-2 antibody STI-9167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Bajic, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | An in vitro experimental pipeline to characterize the epitope of a SARS-CoV-2 neutralizing antibody.
Mbio, 15, 2024
|
|
8EQE
 
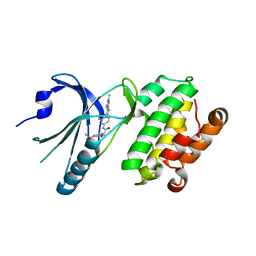 | | Co-crystal structure of PERK with compound 26 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-hydroxy-2-[3-(trifluoromethyl)phenyl]acetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQD
 
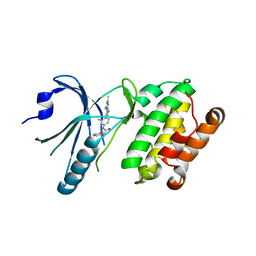 | | Co-crystal structure of PERK with compound 24 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQC
 
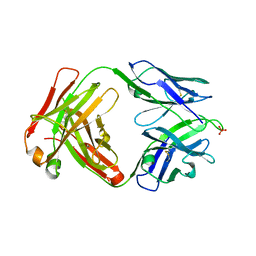 | | Crystal structure of human anti-N1 neuraminidase 3H03 Fab | | Descriptor: | 3H03 Fab heavy chain, 3H03 Fab light chain, PHOSPHATE ION | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8EQA
 
 | | Crystal structure of human anti-N1 neuraminidase 2H08 Fab | | Descriptor: | 2H08 Fab heavy chain, 2H08 Fab light chain, ZINC ION | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8EQ9
 
 | | Co-crystal structure of PERK with compound 11 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQ7
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 20 | | Descriptor: | (2R,3R,4R,5S)-1-[(3-{[3-bromo-5-(methanesulfonyl)anilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EQ1
 
 | |
8EPR
 
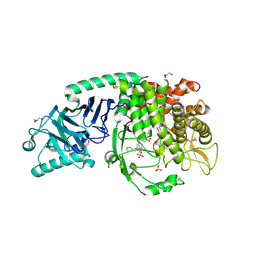 | | Co-crystal structure of Chaetomium glucosidase with compound 19 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[3-({[(5M)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EPO
 
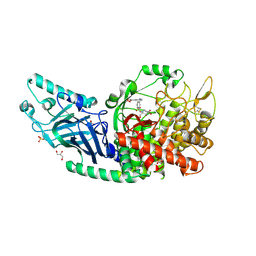 | | Co-crystal structure of Chaetomium glucosidase with compound 18 | | Descriptor: | (3P)-3-(5,6-dihydro-1,4-dioxin-2-yl)-5-{[(3-{[(2R,3R,4R,5S)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidin-1-yl]methyl}phenyl)methyl]amino}benzonitrile, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EPL
 
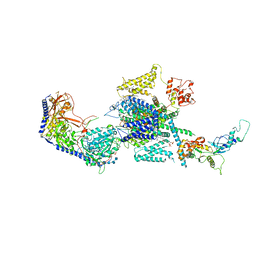 | | Human R-type voltage-gated calcium channel Cav2.3 at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
8EPJ
 
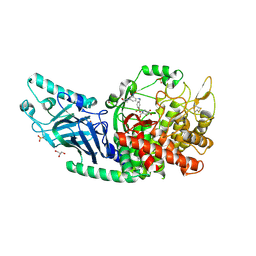 | | Co-crystal structure of Chaetomium glucosidase with compound 17 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[4-(morpholin-4-yl)-2-nitroanilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EPA
 
 | |
8EOP
 
 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EOO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing human antibodies WRAIR-2063 and WRAIR-2151 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Jensen, J.L, Sankhala, R.S, Joyce, M.G. | | Deposit date: | 2022-10-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Targeting the Spike Receptor Binding Domain Class V Cryptic Epitope by an Antibody with Pan-Sarbecovirus Activity.
J.Virol., 97, 2023
|
|
8EOK
 
 | |
8EOC
 
 | | Crystal structure of E.coli DsbA mutant E24A/K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EO5
 
 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EO4
 
 | |
8EO2
 
 | | Lufaxin a bifunctional inhibitor of complement and coagulation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Andersen, J.F, Strayer, E.C. | | Deposit date: | 2022-10-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
8EO1
 
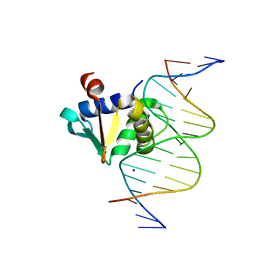 | |
8ENU
 
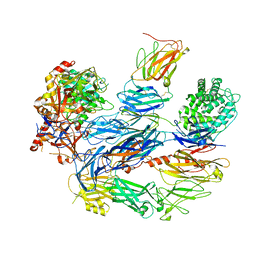 | | Structure of the C3bB proconvertase in complex with lufaxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
8ENR
 
 | |
8ENQ
 
 | | E. coli CsgA fibril (218-pixel box size) | | Descriptor: | Major curlin subunit | | Authors: | Bu, F, Liu, B. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Escherichia coli CsgA amyloid fibril assembly.
Mbio, 15, 2024
|
|
8ENO
 
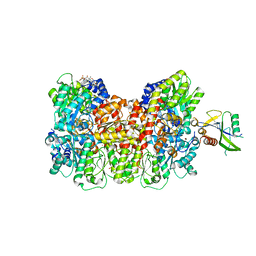 | | Homocitrate-deficient nitrogenase MoFe-protein from A. vinelandii nifV knockout in complex with NafT | | Descriptor: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
